|
|
|
|
|
| Sample: |
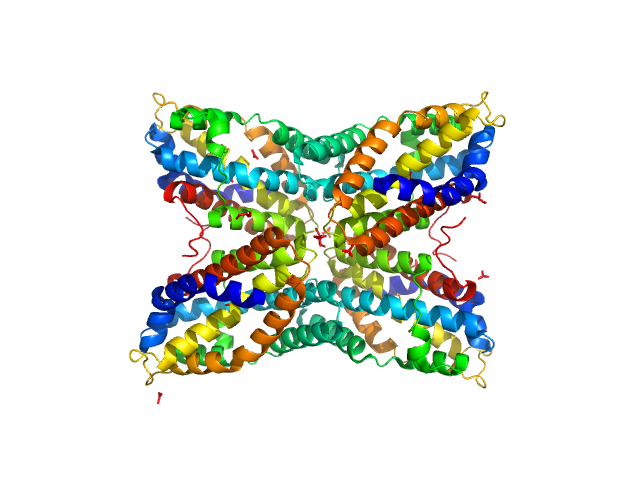
Modification methylase SsoII monomer, 43 kDa Shigella sonnei protein
12-bp DNA monomer, 8 kDa DNA
|
| Buffer: |
50 mM Na-phosphate buffer, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Mar 13
|
Flexibility of the linker between the domains of DNA methyltransferase SsoII revealed by small-angle X-ray scattering: implications for transcription regulation in SsoII restriction-modification syste...
PLoS One 9(4):e93453 (2014)
Konarev PV, Kachalova GS, Ryazanova AY, Kubareva EA, Karyagina AS, Bartunik HD, Svergun DI
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Thiaminase type II enzyme tetramer, 107 kDa Staphylococcus aureus protein
|
| Buffer: |
100 mM Tris-HCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 11
|
Staphylococcus aureus thiaminase II: oligomerization warrants proteolytic protection against serine proteases.
Acta Crystallogr D Biol Crystallogr 69(Pt 12):2320-9 (2013)
Begum A, Drebes J, Kikhney A, Müller IB, Perbandt M, Svergun D, Wrenger C, Betzel C
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - 5α-cholestan-3-one) without Dox monomer, 125 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 19
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
6.1 |
nm |
| Dmax |
20.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - 5α-cholestan-3-one) with Dox (10%) monomer, 220 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 19
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
7.5 |
nm |
| Dmax |
25.5 |
nm |
|
|
|
|
|
|
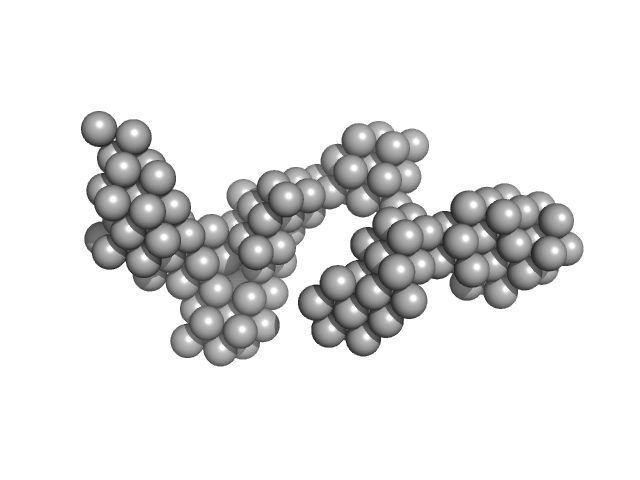
|
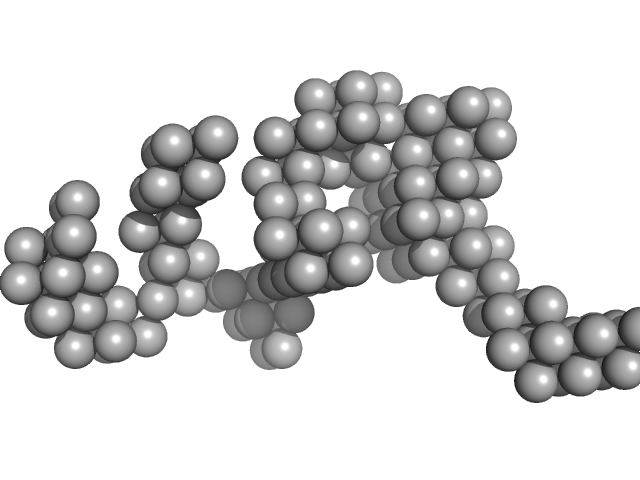
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - Opb-Chol) without Dox monomer, 125 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 19
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
6.4 |
nm |
| Dmax |
22.5 |
nm |
|
|
|
|
|
|
|
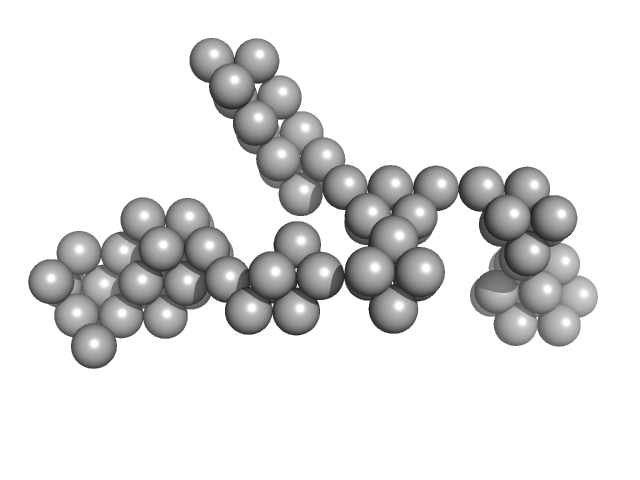
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - Opb-Chol) with Dox (10%) monomer, 225 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 19
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
10.0 |
nm |
| Dmax |
21.9 |
nm |
|
|
|
|
|
|
|
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - Lev-Chol) without Dox monomer, 125 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 19
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
6.5 |
nm |
| Dmax |
22.9 |
nm |
|
|
|
|
|
|
|
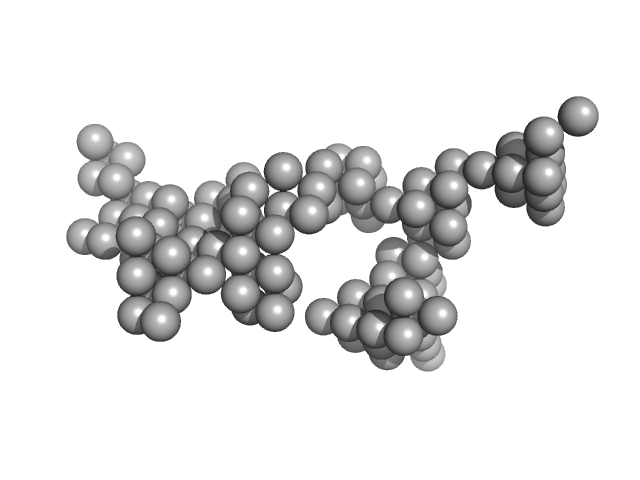
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - Lev-Chol) with Dox (10%) monomer, 125 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 19
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
6.9 |
nm |
| Dmax |
23.8 |
nm |
|
|
|
|
|
|
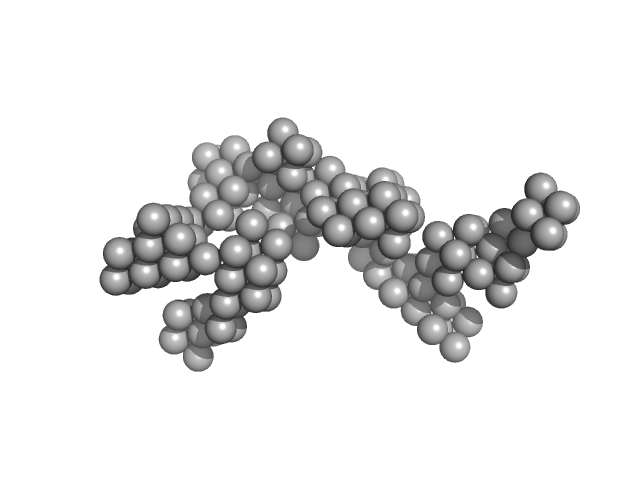
|
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - cholest-4-en-3-one) without Dox monomer, 125 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 16
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
5.9 |
nm |
| Dmax |
19.7 |
nm |
|
|
|
|
|
|
|
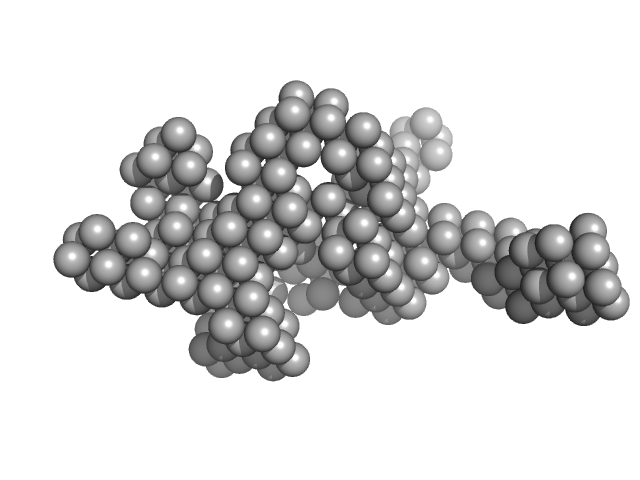
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - cholest-4-en-3-one) with Dox (10%) monomer, 125 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 19
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
5.5 |
nm |
| Dmax |
18.3 |
nm |
|
|