|
|
|
|
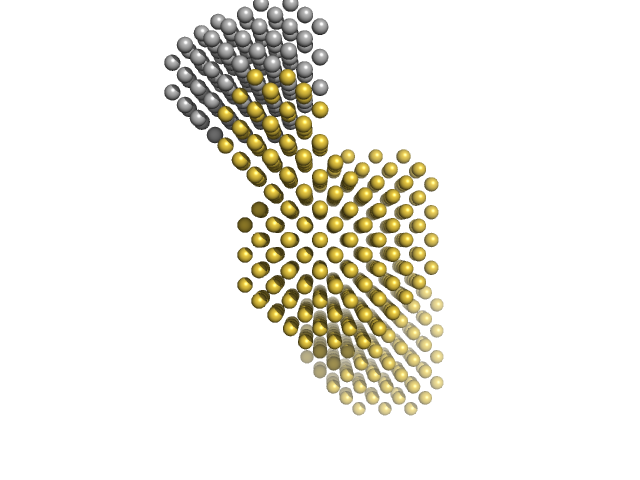
|
| Sample: |
I27-PimA Fusion protein monomer, 48 kDa protein
|
| Buffer: |
50 mM Tris-HCl 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 May 17
|
Conformational plasticity of the essential membrane-associated mannosyltransferase PimA from mycobacteria.
J Biol Chem 288(41):29797-808 (2013)
Giganti D, Alegre-Cebollada J, Urresti S, Albesa-Jové D, Rodrigo-Unzueta A, Comino N, Kachala M, López-Fernández S, Svergun DI, Fernández JM, Guerin ME
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
86 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Estrogen-related receptor gamma hexamer, 230 kDa Homo sapiens protein
Inverse repeat IR3 DNA trimer, 46 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 100 mM KCl, 5 mM MgCl2, 1% (v/v) glycerol, and 1 mM CHAPS, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Mar 3
|
Reconstruction of Quaternary Structure from X-ray Scattering by Equilibrium Mixtures of Biological Macromolecules
Biochemistry 52(39):6844-6855 (2013)
Petoukhov M, Billas I, Takacs M, Graewert M, Moras D, Svergun D
|
| RgGuinier |
4.7 |
nm |
| Dmax |
14.6 |
nm |
|
|
|
|
|
|

|
| Sample: |
Estrogen-related receptor gamma hexamer, 230 kDa Homo sapiens protein
Inverse repeat IR3 DNA trimer, 46 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 100 mM KCl, 5 mM MgCl2, 1% (v/v) glycerol, and 1 mM CHAPS, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Mar 3
|
Reconstruction of Quaternary Structure from X-ray Scattering by Equilibrium Mixtures of Biological Macromolecules
Biochemistry 52(39):6844-6855 (2013)
Petoukhov M, Billas I, Takacs M, Graewert M, Moras D, Svergun D
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.2 |
nm |
|
|
|
|
|
|

|
| Sample: |
Estrogen-related receptor gamma hexamer, 230 kDa Homo sapiens protein
Inverse repeat IR3 DNA trimer, 46 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 100 mM KCl, 5 mM MgCl2, 1% (v/v) glycerol, and 1 mM CHAPS, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Mar 3
|
Reconstruction of Quaternary Structure from X-ray Scattering by Equilibrium Mixtures of Biological Macromolecules
Biochemistry 52(39):6844-6855 (2013)
Petoukhov M, Billas I, Takacs M, Graewert M, Moras D, Svergun D
|
| RgGuinier |
4.1 |
nm |
| Dmax |
12.8 |
nm |
|
|
|
|
|
|

|
| Sample: |
DNA mismatch repair protein MutS dimer, 191 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Feb 28
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
4.7 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
307 |
nm3 |
|
|
|
|
|
|
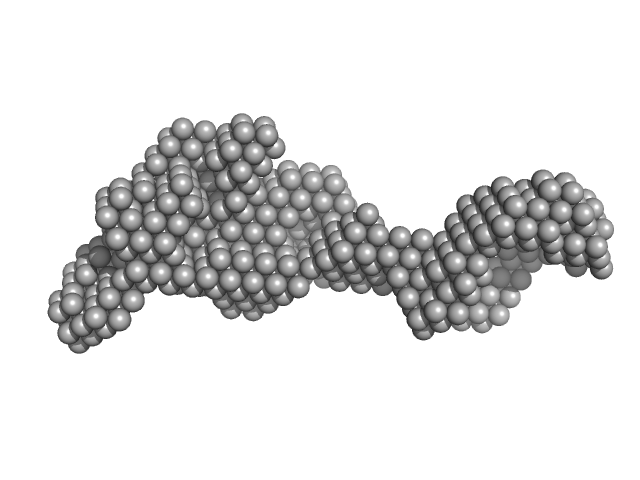
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
7.8 |
nm |
| Dmax |
28.0 |
nm |
| VolumePorod |
700 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
8.5 |
nm |
| Dmax |
29.0 |
nm |
| VolumePorod |
750 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
8.3 |
nm |
| Dmax |
29.0 |
nm |
| VolumePorod |
720 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
|
|
|
|
|
|
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
7.8 |
nm |
| Dmax |
27.0 |
nm |
|
|