|
|
|
|
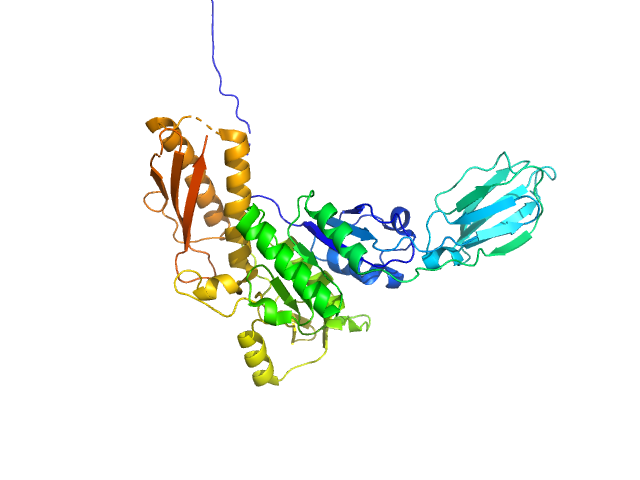
|
| Sample: |
Uncharacterized protein monomer, 48 kDa Desulfitobacterium hafniense protein
|
| Buffer: |
5 mM DTT 100 mM NaCl 10 mM Tris-HCl 0.02 % NaN3, pH: 7.5
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2010 Feb 12
|
Small angle X-ray scattering as a complementary tool for high-throughput structural studies.
Biopolymers 95(8):517-30 (2011)
Grant TD, Luft JR, Wolfley JR, Tsuruta H, Martel A, Montelione GT, Snell EH
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
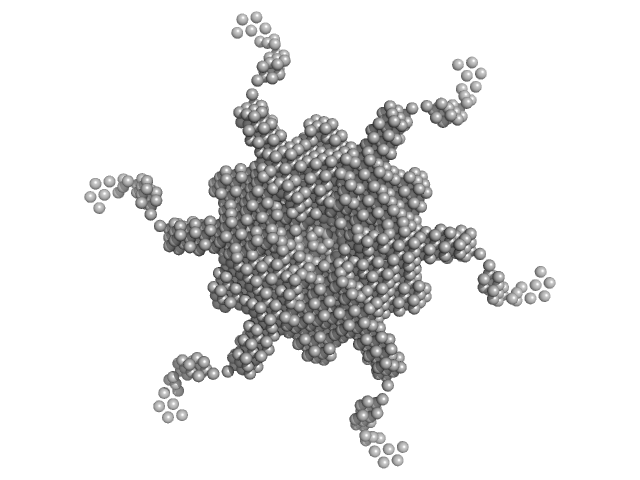
|
| Sample: |
RNA chaperone Hfq hexamer, 67 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 May 2
|
Structural insights into the dynamics and function of the C-terminus of the E. coli RNA chaperone Hfq.
Nucleic Acids Res 39(11):4900-15 (2011)
Beich-Frandsen M, Vecerek B, Konarev PV, Sjöblom B, Kloiber K, Hämmerle H, Rajkowitsch L, Miles AJ, Kontaxis G, Wallace BA, Svergun DI, Konrat R, Bläsi U, Djinovic-Carugo K
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
|
|
|
|
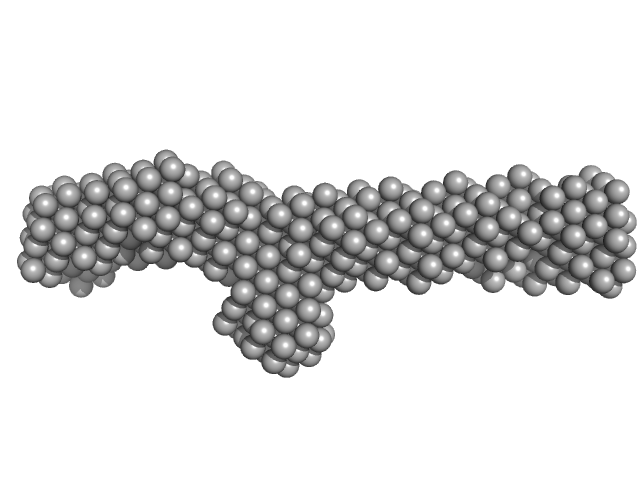
|
| Sample: |
Plectin monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 2.5 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at cSAXS, Swiss Light Source on 2009 Jul 24
|
The structure of the plakin domain of plectin reveals a non-canonical SH3 domain interacting with its fourth spectrin repeat.
J Biol Chem 286(14):12429-38 (2011)
Ortega E, Buey RM, Sonnenberg A, de Pereda JM
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|
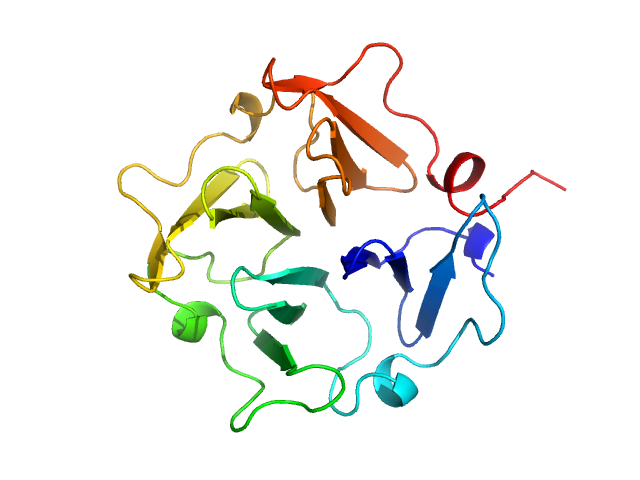
|
| Sample: |
Hemopexin monomer, 23 kDa Homo sapiens protein
Hemopexin dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 10 mM CaCl2, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Nov 25
|
The Dimer Interface of the Membrane Type 1 Matrix Metalloproteinase Hemopexin Domain
Journal of Biological Chemistry 286(9):7587-7600 (2011)
Tochowicz A, Goettig P, Evans R, Visse R, Shitomi Y, Palmisano R, Ito N, Richter K, Maskos K, Franke D, Svergun D, Nagase H, Bode W, Itoh Y
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
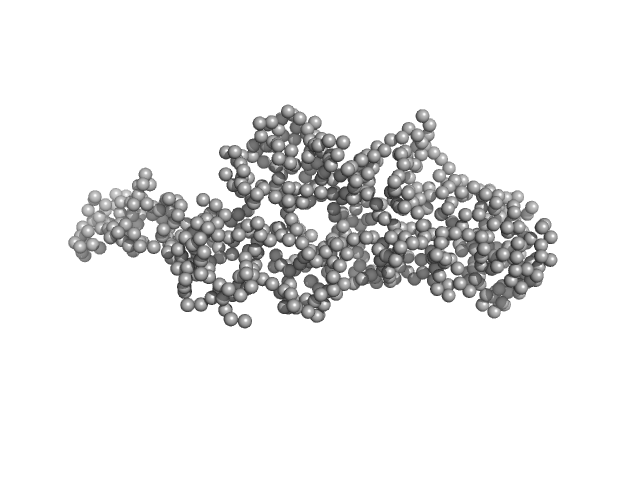
| Sample: |
Multidomain regulatory protein Rv1364c monomer, 70 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
25 mM Tris-HCl, 50 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Feb 14
|
Structural characterization of the multidomain regulatory protein Rv1364c from Mycobacterium tuberculosis.
Structure 19(1):56-69 (2011)
King-Scott J, Konarev PV, Panjikar S, Jordanova R, Svergun DI, Tucker PA
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
|
|
|
|

|
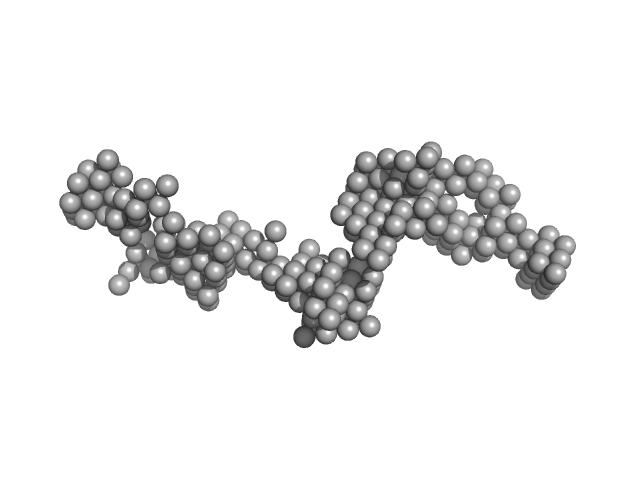
| Sample: |
Multidomain regulatory protein Rv1364c dimer, 139 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
25 mM Tris-HCl, 50 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Feb 14
|
Structural characterization of the multidomain regulatory protein Rv1364c from Mycobacterium tuberculosis.
Structure 19(1):56-69 (2011)
King-Scott J, Konarev PV, Panjikar S, Jordanova R, Svergun DI, Tucker PA
|
| RgGuinier |
4.5 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
289 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein A monomer, 28 kDa Staphylococcus aureus protein
|
| Buffer: |
10 mM HEPES, pH 7.2, 3 mM EDTA, pH: 7.2
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 May 12
|
A structural basis for Staphylococcal complement subversion: X-ray structure of the complement-binding domain of Staphylococcus aureus protein Sbi in complex with ligand C3d
Molecular Immunology 48(4):452-462 (2011)
Clark E, Crennell S, Upadhyay A, Zozulya A, Mackay J, Svergun D, Bagby S, van den Elsen J
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
ImportinA_ImportinB monomer, 160 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Oct 29
|
Recognition of nucleoplasmin by its nuclear transport receptor importin α/β: insights into a complete import complex.
Biochemistry 49(45):9756-69 (2010)
Falces J, Arregi I, Konarev PV, Urbaneja MA, Svergun DI, Taneva SG, Bañuelos S
|
| RgGuinier |
5.7 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
390 |
nm3 |
|
|
|
|
|
|

|
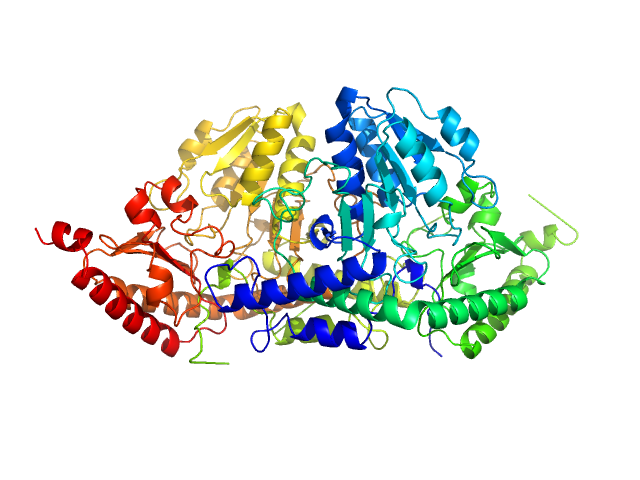
| Sample: |
Nucleoplasmin_importinA_importinB , 900 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 May 23
|
Recognition of nucleoplasmin by its nuclear transport receptor importin α/β: insights into a complete import complex.
Biochemistry 49(45):9756-69 (2010)
Falces J, Arregi I, Konarev PV, Urbaneja MA, Svergun DI, Taneva SG, Bañuelos S
|
| RgGuinier |
8.6 |
nm |
| Dmax |
28.0 |
nm |
| VolumePorod |
1800 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cysteine desulfurase IscS dimer, 87 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl 10 mM β-mercaptoethanol, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 5
|
Structural bases for the interaction of frataxin with the central components of iron-sulphur cluster assembly.
Nat Commun 1:95 (2010)
Prischi F, Konarev PV, Iannuzzi C, Pastore C, Adinolfi S, Martin SR, Svergun DI, Pastore A
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.9 |
nm |
|
|