| MWI(0) | 149 | kDa |
| MWexpected | 173 | kDa |
| VPorod | 234 | nm3 |
|
log I(s)
3.73×103
3.73×102
3.73×101
3.73×100
|
 s, nm-1
s, nm-1
|
|
|
|

|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
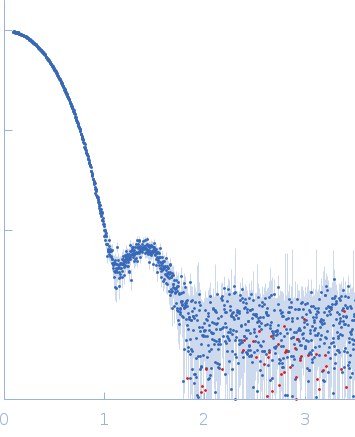
X-ray synchrotron radiation scattering data from solutions of Xylose Isomerase in 20 mM HEPES, 200 mM Na2SO4, 50 mM K2SO4, 50% v/v D2O, 1 mM MgCl2 mM, pH 6.6 were collected on the P12 beam line at Petra-III (Hamburg, Germany) using a Pilatus 2M detector (I(s) vs s, where s = 4π sin θ/λ; 2θ is the scattering angle and λ = 0.12 nm). One solute concentration at 5.76 mg/ml was measured using a total exposure time of 1 second (20 x 0.050 second frames). The data were normalized to the intensity of the transmitted beam and radially averaged. Scattering contributions from the matched solvent blank were subtracted and the resulting 1D scattering data were scaled to protein concentration. The data presented in this entry were obtained from a single concentration scattering curve.
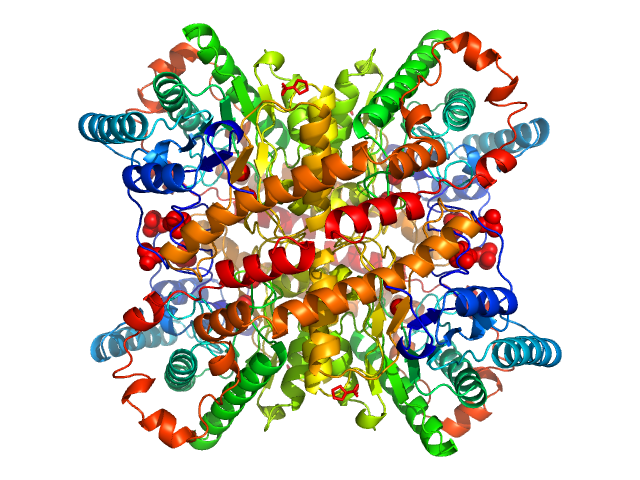
The data displayed at the top of the entry, and the subsequent ab inito and atomic model fit, are data derived from averaging 20 individual frames. The individual 20 data frames used for the averaging are also displayed, with the respective atomic-model fits (CRYSOL; PDB 4E3V).
Tags:
benchmark
|
|
|||||||||||||||||||||||||||