|
|
|
|
|
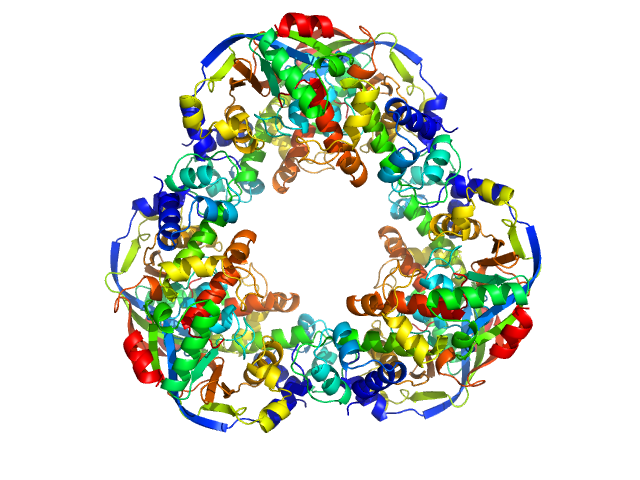
| Sample: |
Mycobacterial cidal toxin hexamer, 121 kDa Mycobacterium tuberculosis protein
Mycobacterial cidal antitoxin hexamer, 76 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
100 mM HEPES, 100 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
An NAD+ Phosphorylase Toxin Triggers Mycobacterium tuberculosis Cell Death.
Mol Cell (2019)
...Svergun DI, Schnappinger D, Schneider TR, Genevaux P, de Carvalho LPS, Wilmanns M, Parret AHA, Neyrolles O
|
| RgGuinier |
4.1 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
262 |
nm3 |
|