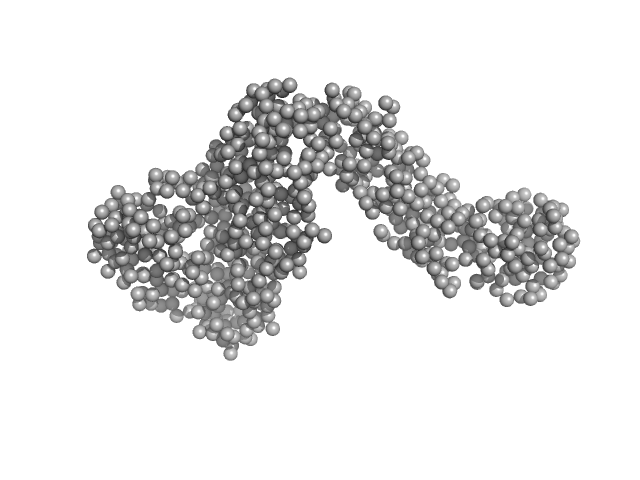|
Synchrotron SAXS data from solutions of Shigella flexneri outer membrane protein IcsA (53–758) in 50 mM Tris, 150 mM NaCl, 10 mM CaCl2, 3% v/v glycerol, pH 7.4 were collected using size-exclusion chromatography (SEC) SAXS on the EMBL-P12 bioSAXS beam line at the PETRAIII storage ring (Hamburg, Germany) equipped with a Pilatus 2M detector (I(s) vs s, where s = 4π sin θ/λ; 2θ is the scattering angle; λ = 0.124 nm). 119 successive 1 second frames were collected through the monomeric IcsA SEC elution peak (GE Healthcare Life Sciences Superdex 200 Increase column; 0.4 ml/min; 75 μl injection at 8 mg/ml). The data were normalised to the intensity of the transmitted beam and the scattering of the solvent-blank (derived from 50 x 1 s data frames of pure, macromolecular free solvent) was subtracted. The resulting scattering profiles were scaled relative to each other (CorMap p > 0.01) then averaged to produce the final averaged IcsA SAXS data displayed in this entry.
The bead models displayed for IcsA are individual/representative low-resolution structural examples calculated using DAMMIN or GASBOR. A cohort of individual GASBOR and DAMMIN models are supplied in the full entry zip archive and include the average DAMMIN-based spatial representation of IcsA in solution (damfilt.pdb; NSD = 0.7).
Individual reduced SEC-SAXS data frames/blocks and Rg/I(0) analysis through the IcsA elution peak are included in the full entry zip archive.
|
|
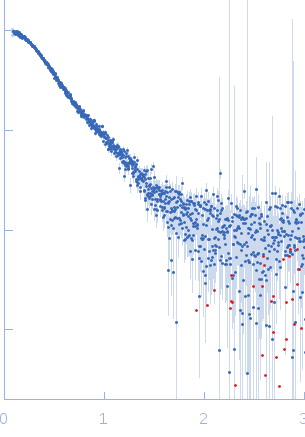 s, nm-1
s, nm-1