|
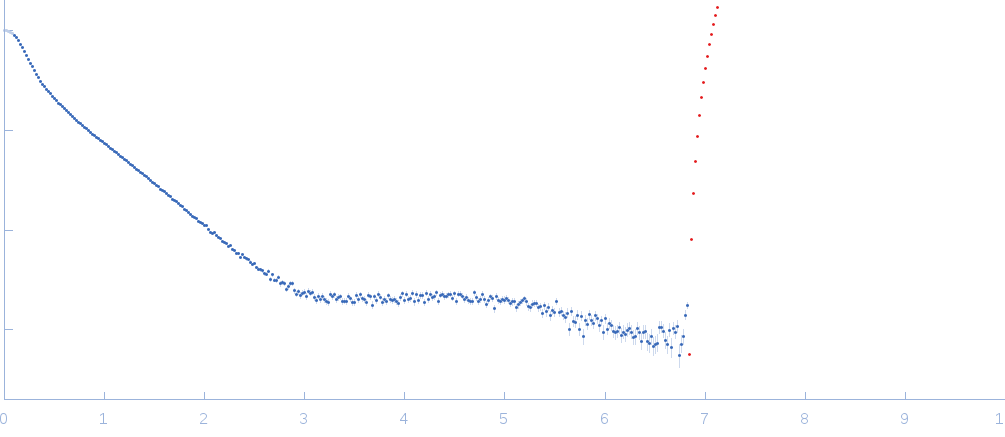
SAXS data from solutions of Domain 3'X of hepatitis C virus RNA at higher ionic strength in 10mM Tris 0.1 mM EDTA 2 mM MgCl2 50 mM NaCl, pH 7, were collected on an in-house Rigaku BIOSAXS-2000 instrument (Frederick, USA) using a Pilatus 100K detector at a sample-detector distance of 0.5 m and at a wavelength of λ = 0.154 nm (I(s) vs s, where s = 4π sin θ/λ and 2θ is the scattering angle). Solute concentrations ranging between 0.4 and 1.5 mg/ml were measured at 27°C. 30 successive 1 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted and the different curves were scaled for RNA concentration. The low angle data collected at lower concentrations were extrapolated to infinite dilution and merged with the higher concentration data to yield the final composite scattering curve.
|
|
 s, nm-1
s, nm-1
