|
Synchrotron SAXS data from solutions of the catalytic domain (AC) of B. Pertussis adenylate cyclase toxin (CyaA) in 20 mM HEPES, 150 mM NaCl, 4 mM CaCl2, pH 7.4 were collected on the SWING beam line at the SOLEIL storage ring (Saint-Aubin, France) using a CCD AVIEX detector at a sample-detector distance of 2.0 m and at a wavelength of λ = 0.1 nm (I(s) vs s, where s = 4πsin θ/λ and 2θ is the scattering angle). 250 successive 1.500 second frames were collected at at 15°C using size-exclusion chromatography SAXS.
The scattered intensities were displayed on an absolute scale (cm-1) using the scattering of water. Frames were examined individually and 20 identical frames were averaged and further processed. The corresponding concentration was 0.80 g/L.
Three independent determinations of the molecular mass were obtained from the value of I(0)/c, where c is the protein concentration, and using the programs SAXSMow2 and ScÅtter3 available at the URLs http://saxs.ifsc.usp.br/ and https://bl1231.als.lbl.gov/scatter/, respectively. The average value is MWexperimental=40.2 ± 1.0 kDa.
AC in solution:
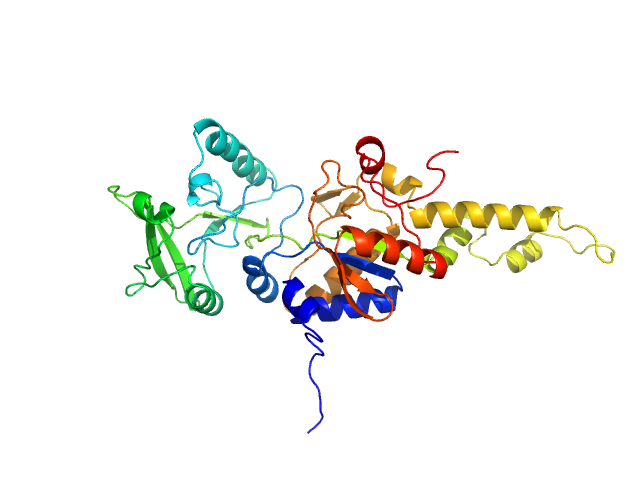
Top panel: Adjustment of the curve calculated from the crystal structure of AC extracted from the pdb dataset 1YRU (red curve) after missing loop addition using AllosModFoxs against experimental data (blue dots). chi2=2.57
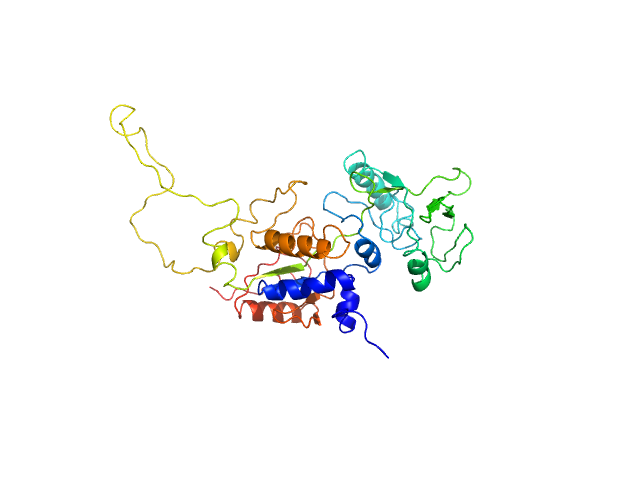
Bottom panel: Adjustment obtained using AllosModFoxs by releasing residues 200-270 comprising helices F through H’ (red curve). Residues 200-270 correspond to the large yellow-light green loop seen in the model displayed on the right. chi2=1.36
|
|
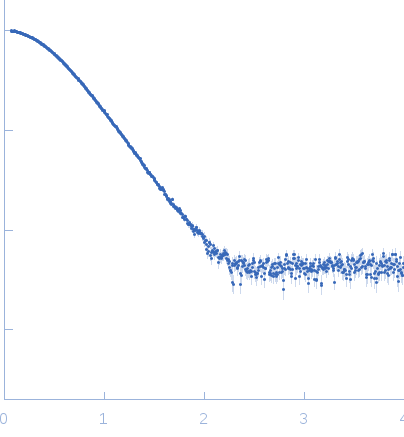 s, nm-1
s, nm-1