| MWI(0) | 178 | kDa |
| MWexpected | 200 | kDa |
| VPorod | 415 | nm3 |
|
log I(s)
1.97×100
1.97×10-1
1.97×10-2
1.97×10-3
|
 s, nm-1
s, nm-1
|
|
|
|
|
|

|
|
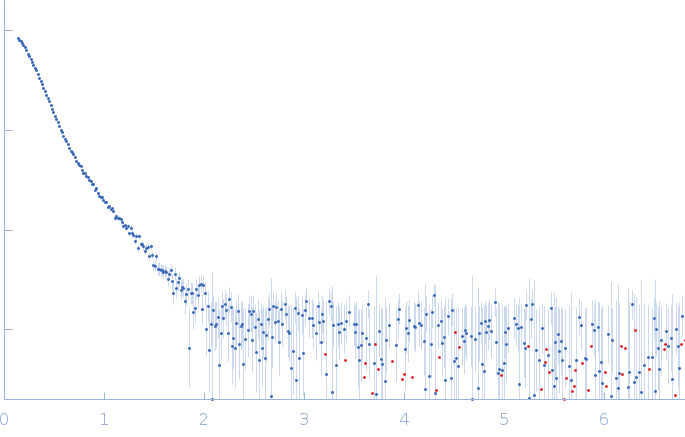
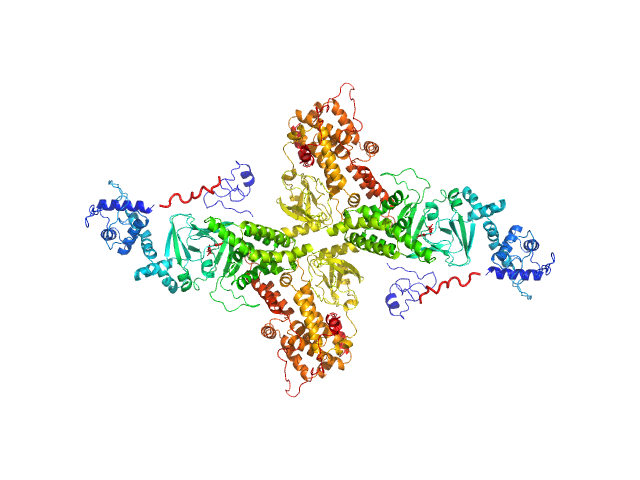
SAXS data from solutions of Rap guanine nucleotide exchange factor 3 (isoform3) - binary form with cAMP - in 1mM EDTA, 10mM DTT, 500mM NaCl, 1mM cAMP, 10mM Tris, pH 9 were collected using a Rigaku BioSAXS-1000 instrument at the Sealy Center For Structural Biology, UTMB-G (Galveston, TX, USA) equipped with a Kratky-2D Rigaku BioSAXS-1000 detector at a sample-detector distance of 0.5 m and at a wavelength of λ = 0.15418 nm (I(s) vs s, where s = 4πsin θ/λ and 2θ is the scattering angle). A sample concentration of 4 mg/ml was measured at 10°C. SAXS data were acquired from 4 successive 1800 second frames. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted to generate the data displayed for this entry.
|
|
|||||||||||||||||||||||||||||||||