|
SAXS experiments were performed at the SAXS2 beamline in the Laboratório Nacional de Luz Síncrotron (LNLS, Campinas-SP, Brazil). The X-ray scattering data (I(s) vs s, where s = 4πsinθ/λ and 2θ is the scattering angle; λ = 0.1488 nm) were acquired using a two-dimensional MAR-CCD detector. Measurements were performed with a monochromatic X-ray beam and a sample-to-detector distance of ~1000 mm, corresponding to a scattering vector range of 0.015 < q < 0.35 Å−1. Scattering patterns were recorded at different sample concentrations (approximately 1.0, 2.0 and 4.0 mg/mL) and several frames in order to check for protein aggregation and X-ray damage. No evidence of aggregation or inter-particle interference were observed.
Number of frames = UNKNOWN
|
|
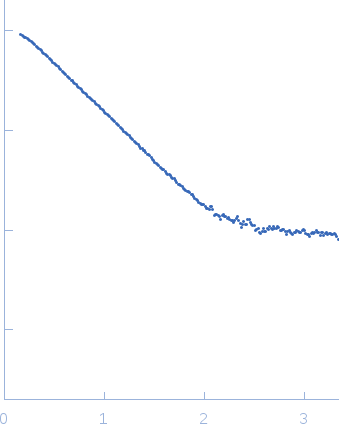 s, nm-1
s, nm-1
