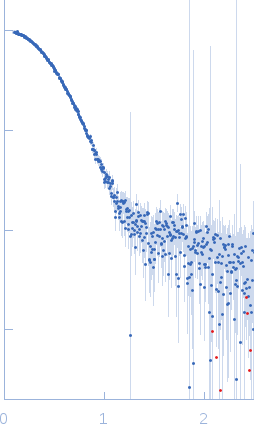
| MWI(0) | 86 | kDa |
| MWexpected | 76 | kDa |
| VPorod | 217 | nm3 |
|
log I(s)
2.51×101
2.51×100
2.51×10-1
2.51×10-2
|
 s, nm-1
s, nm-1
|
|
|
|
|
|

|
|
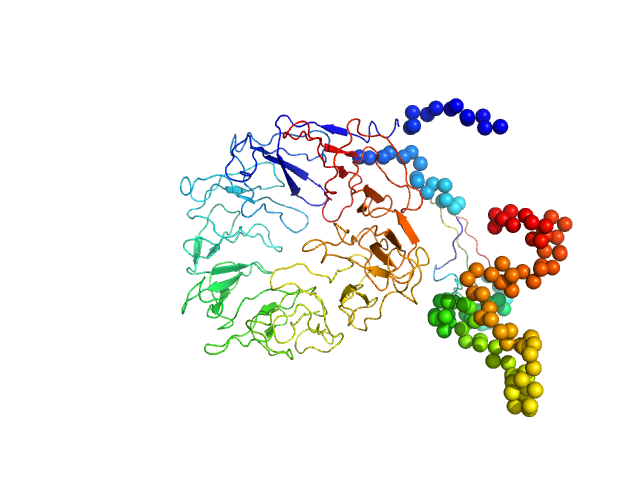
Synchrotron SAXS data from solutions of monomeric sortilin at pH 7.4 in 25 mM HEPES, 150 mM NaCl, pH 7.4 were collected on the BM29 beam line at the ESRF (Grenoble, France) using a Pilatus 1M detector at a sample-detector distance of 2.9 m and at a wavelength of λ = 0.099 nm (I(s) vs s, where s = 4πsinθ/λ and 2θ is the scattering angle). Size-exclusion chromatography SAXS (SEC-SAXS) was performed at 20°C where 2600 successive 1 second frames were collected through the elution profile. The data were normalized to the intensity of the transmitted beam and radially averaged and the scattering of an appropriate solvent-blank was subtracted from the SAXS data measured from the sample component elution peaks.
Concentration min = UNKNOWN |
|
|||||||||||||||||||||||||||