|
Synchrotron SAXS data from solutions of the LIM domains of LIM/homeobox protein Lhx4 fused to the LIM interaction domain (LID; R282G mutant) of the insulin gene enhancer protein ISL-2 in 20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH 8 were collected on the SAXS/WAXS beam line at the Australian Synchrotron (Melbourne, Australia) using a Pilatus 1M detector at a wavelength of λ = 0.10332 nm (l(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). Solute concentrations ranging between 0.2 and 1.9 mg/ml were measured at 13.5°C. 24 successive 1 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted. The low angle data collected at lower concentrations were extrapolated to infinite dilution and merged with the higher concentration data to yield the final composite scattering curve.
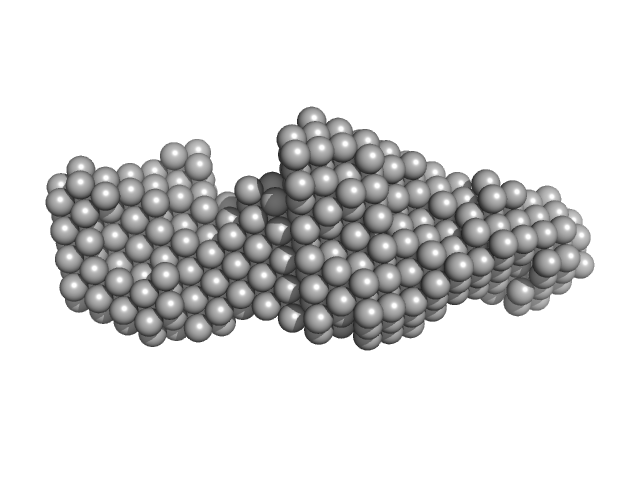
The protein anaysed is a fusion of two interacting partners: the LIM domains of Lhx4 and the LID of Isl2, with a point mutation within the Isl2-LID (R282G). The ab initio bead models presented in this entry include: 1) The best-fit individual reconstruction (top) and; 2) The spatially aligned and volume/bead occupancy-corrected averaged representation of the protein in solution (bottom) calculated from twenty individual reconstructions (NSD = 0.67; resolution estimate = 2.8 nm). Five concentrations of protein were subjected to SAXS as static samples (96-well plate) at the Australian Synchrotron. Subtle concentration dependent effects were noted. All data and additional information relating to the concentration series is included in the full-entry zip archive. The protein was subjected to SEC (off line) as the last purification step just prior to data collection. Also refer to the related SASBDB entry SASDDQ4.
|
|
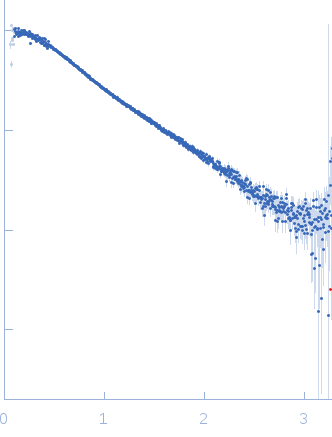 s, nm-1
s, nm-1