|
Synchrotron SAXS data from solutions of the DacA in 30 mM Tris, 150 mM NaCl, pH 7.5 were collected using size-exclusion chromatography SAXS (SEC-SAXS) on the B21 beam line at the Diamond Light Source (Oxfordshire, UK) using a Pilatus 2M detector at a sample-to-detector distance of 4.014 m and X-ray wavelength of λ = 0.1 nm (l(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). SEC-SAXS was performed at 20°C using the following parameters: Column: Superdex 200 5/150 (GE Healthcare); Flow rate: 0.075 mL/min; Sample injection concentration: 12.0 mg/mL; Injection volume: 45 μL. The data obtained through the sample elution peak (collected as consecutive 3 s exposures) were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted and the individual subtracted data sets were scaled and averaged to generate the scattering profile displayed in this entry.
Number of frames = UNKNOWN. SEC column = UNKNOWN. Sample injection volume = UNKNOWN. Flow rate = UNKNOWN
|
|
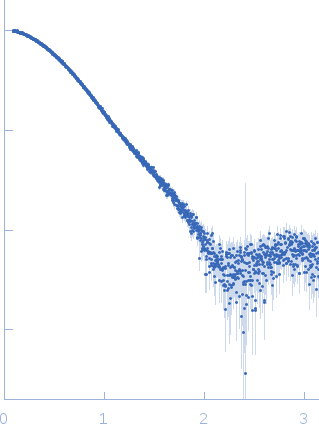 s, nm-1
s, nm-1
