|
Synchrotron SAXS data, I(s) vs s (where s = 4πsinθ/λ, and 2θ is the scattering angle), were measured from the C-terminal half of the Pseudorabiesvirus tegument protein UL37 using continuous-flow size-exclusion chromatography SAXS (SEC-SAXS) on the G1 beam line at the Cornell High Energy Synchrotron Source (CHESS; Ithaca, NY, USA). Data were collected a using a Pilatus 200K detector at a sample-detector distance of 1.5 m and at a wavelength of λ = 0.125 nm. The SEC mobile phase consisted of 100 mM HEPES, 150 mM NaCl, 5% glycerol, 0.1 mM tris(2-carboxyethyl)phosphine (TCEP), pH 7.5, (4°C). The SAXS data measured from the SEC-elution (sample peak and buffer; 300 successive 2 s frames) were normalized to the intensity of the transmitted beam and radially averaged. The scattering of an appropriate solvent-blank was subtracted from the sample frames to produce the scaled and averaged data displayed in this entry.
SEC column = UNKNOWN. Sample injection volume = UNKNOWN. Flow rate = UNKNOWN
|
|
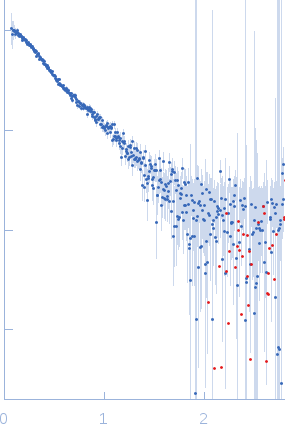 s, nm-1
s, nm-1