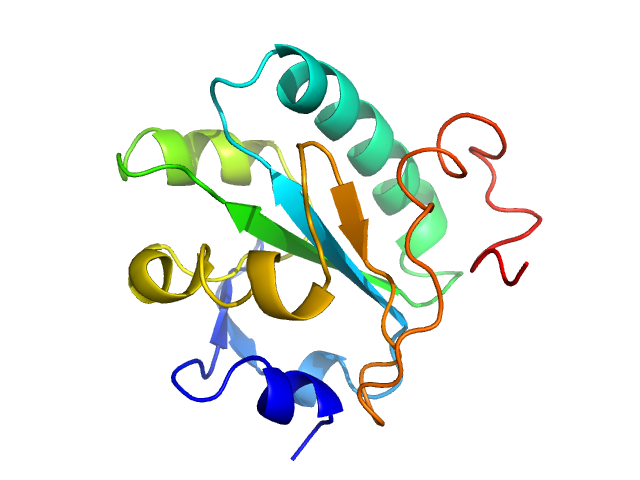
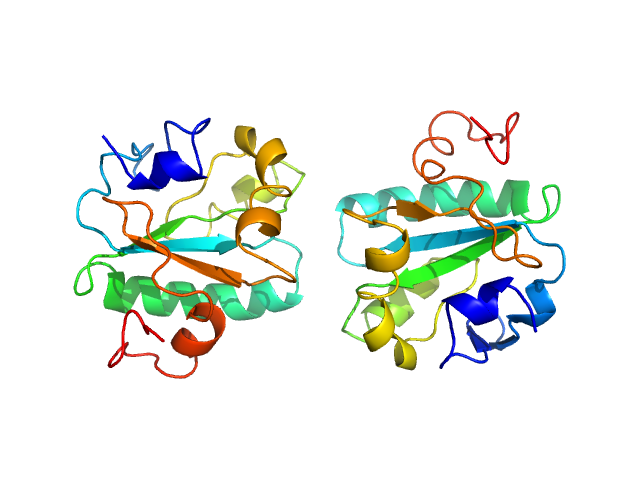
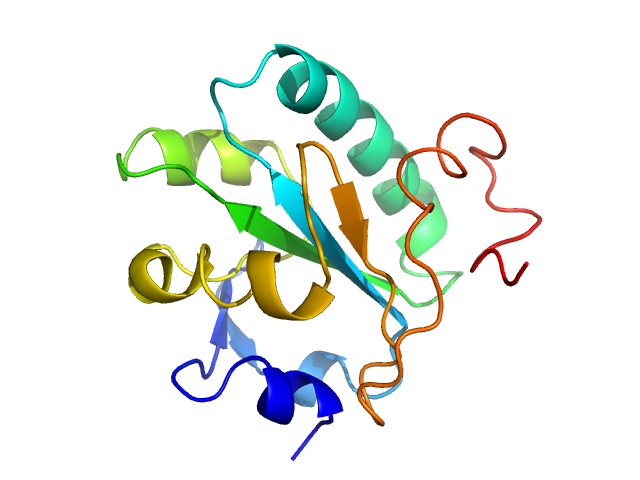
| MWexperimental | 16 | kDa |
| MWexpected | 16 | kDa |
| VPorod | 27 | nm3 |
|
log I(s)
1.00×101
1.00×100
1.00×10-1
1.00×10-2
|
 s, nm-1
s, nm-1
|
|
|
|
|
|
|
|
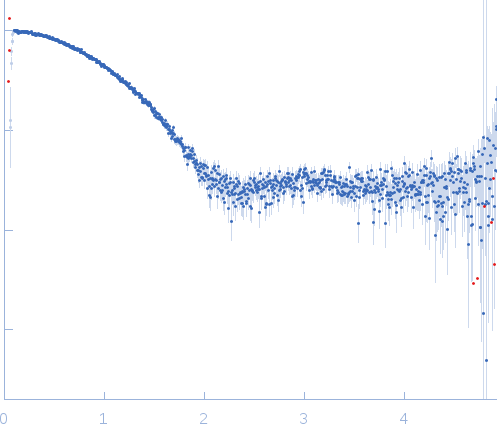
Synchrotron SAXS data from solutions of Tryparedoxin, reduced state, in 10 mM HEPES pH 7.5, 50 mM NaCl, pH 7.5 were collected on the BM29 beam line at the ESRF (Grenoble, France) using a Dectris Pilatus 1M detector at a sample-detector distance of 2.9 m and at a wavelength of λ = 0.099 nm (l(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). The data were collected using in-line size exclusion chromatography (SEC)-SAXS at 20°C.
SEC column type: GE Healthcare S75 3.2/300; Sample Injection Concentration = 10 mg/mL; Column flow-rate = 0.1 mL/min. |
|
|||||||||||||||||||||||||||||||||||||||||||||