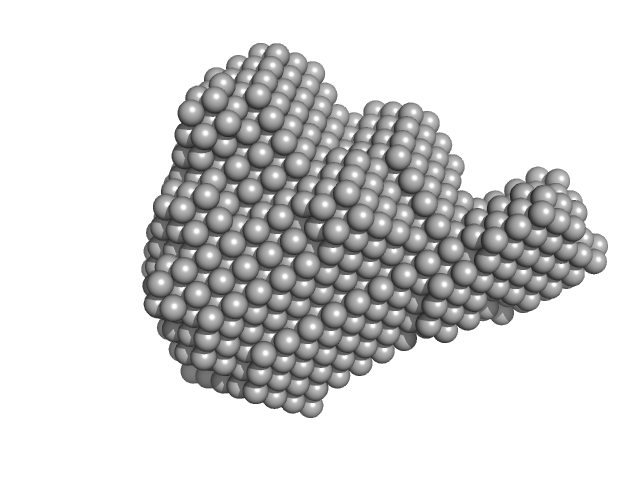
|
Synchrotron SAXS data from solutions of the H2A:H2B:H3:H4 histone complex bound to the acidic domain of APLF in 25 mM NaPi, 300 mM NaCl, 3% v/v glycerol, 1 mM DTT, pH 7 were collected on the EMBL P12 beam line at PETRA III (DESY, Hamburg, Germany) using a Pilatus 6M detector at a sample-detector distance of 3 m and at a wavelength of λ = 0.12 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). 40 successive 1 second frames were collected from a sample and buffer at 20°C. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted. The low angle data collected at lower concentration were merged with the highest concentration high angle data to yield the final composite scattering curve.
The data show the merged SAXS data obtained from batch and SEC-UV-SAXS measurements and therefore the concentration is not specified. The SEC parameters were: Column type: GE Superdex S200 Increase 10/300. Flow rate: 0.6 ml/min. Injection volume and load concentration: 40 µl at 14 mg/mL. Total run time: 35 min. NOTE: The complex consists of one histone complex (8 subunits) and two bound APLF molecules. However, the APLF molecules are not in contact with each other and are a separated on the histone complex. The molecular weight of the complex was validated using mass spectrometry. The molecular weight, from native mass-spec gives 122,926.1 +/- 9.6 Da, for expected mass of 122,839.8 Da based on the atomic composition.
|
|
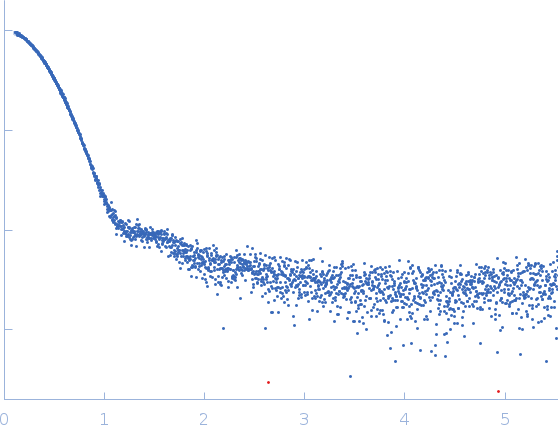 s, nm-1
s, nm-1