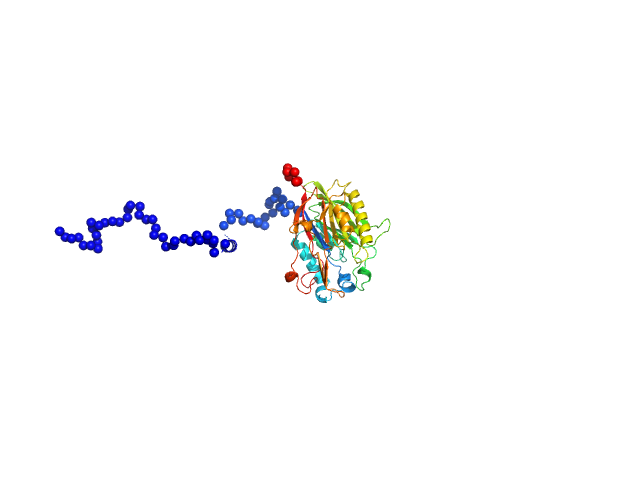
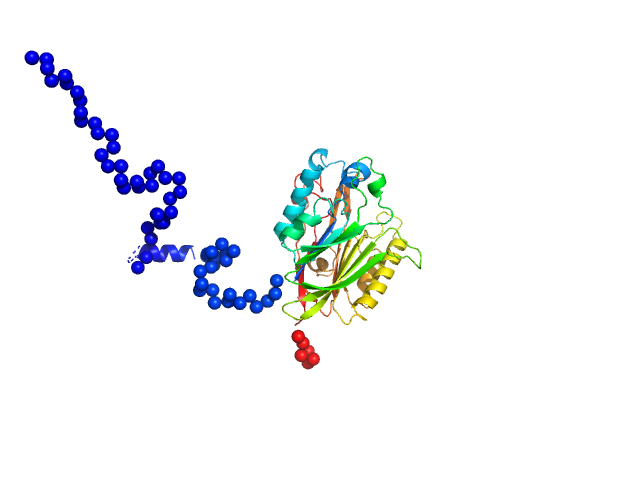
|
SAXS was performed at BioCAT (beamline 18ID at the Advanced Photon Source, Chicago) with in-line size exclusion chromatography (SEC) to separate sample from aggregates and other contaminants thus ensuring optimal sample quality and multiangle light scattering (MALS), dynamic light scattering (DLS) and refractive index measurement(RI)) for additional biophysical characterization (SEC-MALS-SAXS). The samples were loaded on a Superdex 200 Increase 10/300 GL column (Cytiva) run by a 1260 Infinity II HPLC (Agilent Technologies) at 0.5 ml/min. The flow passed through (in order) the Agilent UV detector, a MALS detector and a DLS detector (DAWN Helios II, Wyatt Technologies), and an RI detector (Optilab T-rEX, Wyatt). The flow then went through the SAXS flow cell. The flow cell consists of a 1.0 mm ID quartz capillary with ~20 µm walls. A coflowing buffer sheath is used to separate sample from the capillary walls, helping prevent radiation damage (Kirby et al., 2016). Scattering intensity was recorded using a Pilatus3 X 1M (Dectris) detector which was placed 3.63 m from the sample giving us access to a s-range of 0.042 to 3.5 nm-1. 0.5 s exposures were acquired every 1 s during elution and data was reduced using BioXTAS RAW 2.1.0 (Hopkins et al., 2017). Buffer blanks were created by averaging regions flanking the elution peak and subtracted from all the exposures in the dataset. Molecular weights and hydrodynamic radii were calculated from the MALS and DLS data respectively using the ASTRA 7 software (Wyatt).
After buffer subtraction, both the wild type and LZ data were found to contain multiple scattering components in the elution peak that were not separated by the size exclusion step. Evolving factor analysis (EFA) (Meisburger et al., 2016) as implemented in RAW was used to deconvolve the overlapping data into subtracted scattering profiles for each species. This allowed direct analysis of the species independently. For the 118-NOCT data, the subtracted scattering profile was obtained by averaging a range of data across the elution peak using RAW. Further data analysis including: determination of the radius of gyration (Rg) and forward scattering (I(0)) via Guinier analysis; molecular weight calculation by adjusted Porod volume (Piiadov et al., 2018), volume of correlation (Rambo & Tainer, 2013), DATCLASS (Franke et al., 2018), and the Baseyian inferrance method of DATMW (Hajizadeh et al., 2018); dimensionless Kratky plots; and calculation of the pair distance distribution function (P(r)) using the indirect Fourier transformation method of GNOM (Svergun, 1992) was done in RAW. DATCLASS, DATMW, and GNOM are programs from the ATSAS 3.0.3 package (Manalastas-Cantos et al., 2021) with GUIs implemented in RAW.
|
|
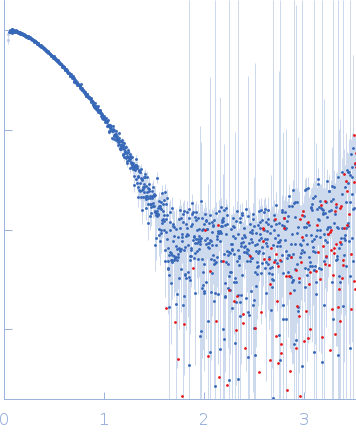 s, nm-1
s, nm-1
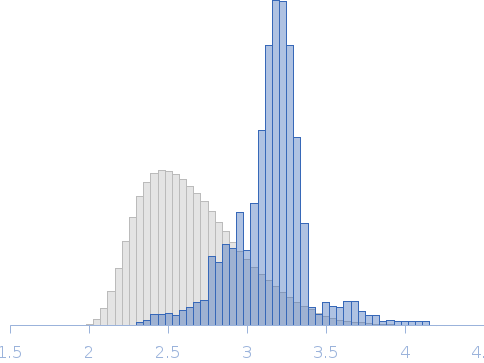 Rg, nm
Rg, nm