|
SAXS data from solutions of the hepatitis C virus RNA Domain 3'X mutant/subdomain 5BSL3.2 complex in 10 mM Tris-HCl, 6 mM MgCl2, pH 7 were collected using a Rigaku BioSAXS-1000 instrument at the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK; National Institutes of Health, Bethesda, MD, USA) equipped with a Pilatus 100K detector at a sample-detector distance of 0.5 m and at a wavelength of λ = 0.154 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). Solute concentrations ranging between 0.5 and 1.5 mg/ml were measured at 27°C. Eight successive 1 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted. The low angle data collected at lower concentrations were extrapolated to infinite dilution and merged with the higher concentration data to yield the final composite scattering curve.
The bead models presented in this entry include: 1) Top - The spatially aligned and volume/bead occupancy-corrected averaged representation calculated from the spatial alignment of a cohort of individual model reconstructions. 2) Bottom - An individual model reconstruction and associated individual model fit to the data.
|
|
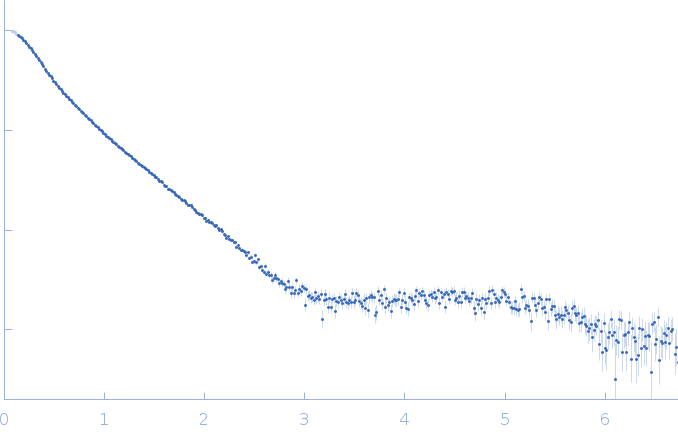 s, nm-1
s, nm-1

