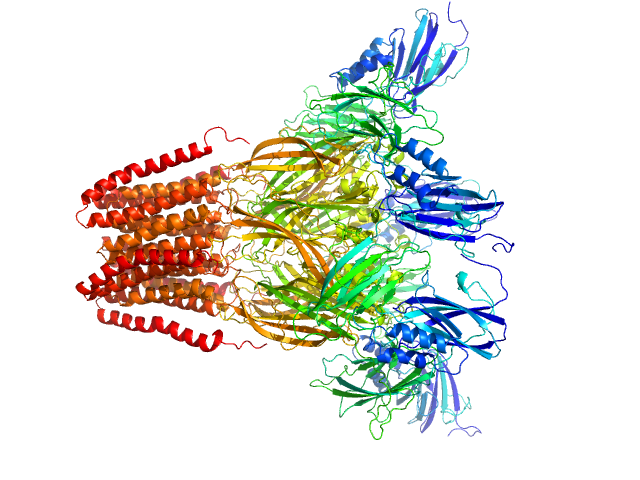
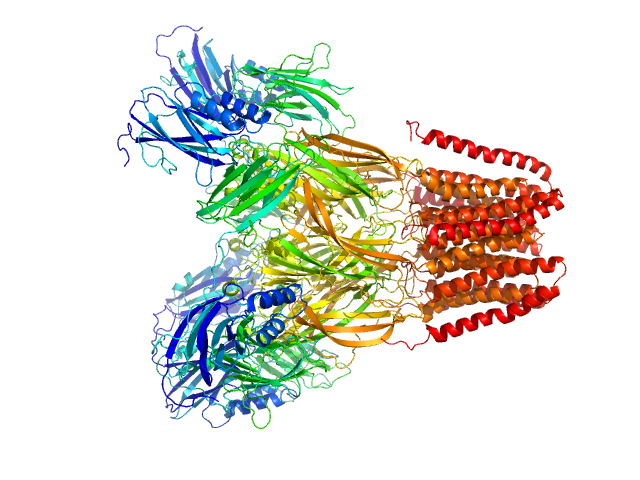
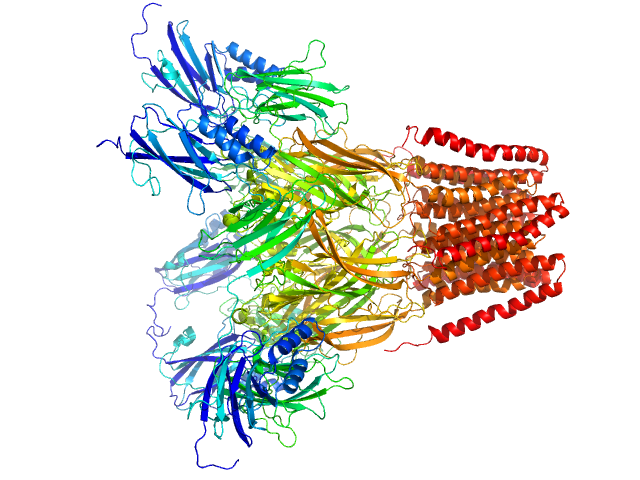
|
SANS data from solutions of Desulfofustis sp. PB-SRB1 ligand-gated ion channel (DeCLIC) at pH 7.5 measured with paused-flow SEC-SANS in D2O, 20 mM Tris, 150 mM NaCl, 10 mM CaCl2, and 0.5 mM matched-out deuterated DDM, were collected on the D22 small-angle neutron scattering diffractometer at the Institut Laue-Langevin (ILL; Grenoble, France) using a Reuter-Stokes 3He detector at a wavelength of λ = 0.6 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). The ion channel DeCLIC was expressed in Escherichia coli and purified using H2O based buffers and n-Dodecyl β-D-maltoside (DDM) detergent. The protein was exchanged to the D2O based buffer with deuterated DDM (d-DDM) in the SEC-SANS gel filtration. The d-DDM has been deuterated to have the same scattering length density as D2O (matched-out deuteration). The gel filtration was run with a 0.2 ml/min flow speed to allow for detergent exchange prior to the SANS measurement. Upon peak detection by UV-vis absorbance at 280 nm the flow was slowed to 0.01 ml/min, and fully paused to 0 ml/min at the peak max. Prior and post the pausing of the flow SANS data was collected with a detector distance of 2.8 m, while paused SANS data was collected with a detector distance of 8 m. The flow was restarted to 0.01 ml/min for the remainder of the peak, after which the flow was increased to pass the remainder of the column volume. Data reduction and buffer subtraction were performed using GRASP version 9.04, utilizing a dedicated buffer measurements for the buffer background. Data were corrected for the empty cuvette and background, and scaled by their transmission and thickness. They were scaled to absolute intensity by direct flux measurement. Concentration normalization, bringing the intensity unit to 1/cm ml/mg, was performed by dividing the scattering intensity with the concentration calculated using the absorbance at 280 nm from the co-recorded chromatograms and the extinction coefficient calculated from the amino acid sequence using ProtParam. In merging the data from the two detector distances, the data from 8 m was used up to 0.9 1/nm, above which the data from 2.8 m was used. A small additional constant was subtracted as a final adjustment to the background. Guinier analysis was performed and the molecular weight estimated from I(0). The pair-distance distribution was calculated using BayesApp (https://somo.chem.utk.edu/bayesapp/), set to fit the background and with no specified maximum diameter. Atomic models were generated from crystal structures (6v4a and 6v4s) and cryo-EM structures (7q3g and 7q3h) of DeCLIC, followed by molecular dynamics simulations in GROMACS. Fits of atomic models to the experimental SANS data was performed using Pepsi-SANS (v. 3.0). The three models with the best fit to the SANS data, which all were from simulations of 7q3h, have been deposited. In the experimental data the uncertainty on the estimated s-value (sigma s) is reported in the fourth column in addition to the error for the intensity (the third column). Fits in Pepsi-SANS were run with all four columns of experimental data, but in the submitted model fits the sigma s column has been removed.
Storage temperature = UNKNOWN
|
|
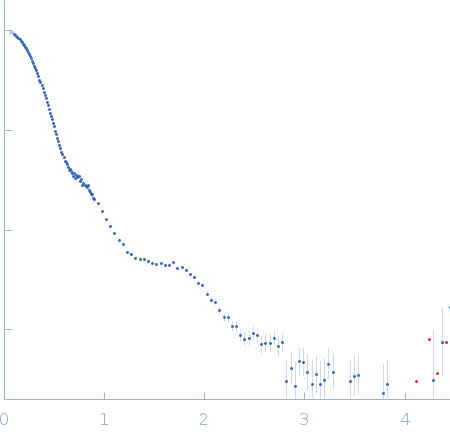 s, nm-1
s, nm-1