|
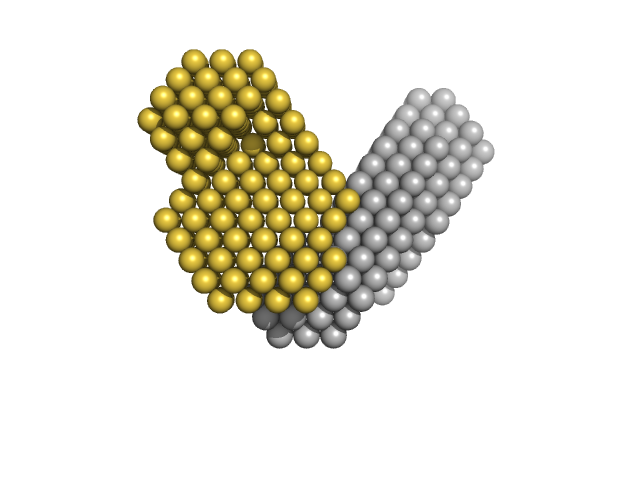
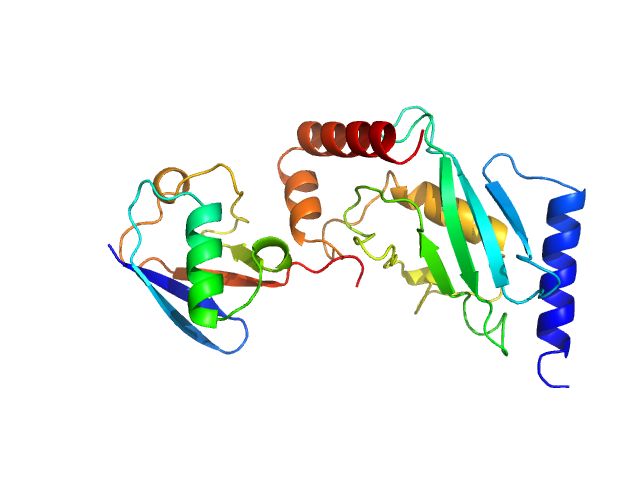
SANS data from solutions of ubiquitin-conjugating enzyme E2 conjugated to deuterated ubiquitin in 50 mM Tris, 150 mM NaCl, 1 mM TCEP, pD 8, were collected on the Quokka small angle neutron scattering instrument at the Australian Centre for Neutron Scattering (ANSTO, Lucas Heights, Australia) using a ORDELA/21000N detector at a wavelength of λ = 0.6 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). One solute concentration of 4.02 mg/ml was measured at 10°C. Contrast variation SANS data (made available in the full entry zip archive) were collected at two sample-to-detector positions, 6 m (long configuration, 0.1 < s < 1.1 1/nm) and 1.3 m (short configuration, 0.5 < s < 4.8 1/nm), that were subsequently merged to generate the final SANS profiles. The neutron wavelength spread was 10%. The protein conjugate in 0% v/v D2O buffer was measured for 3,600 s (long configuration) and 3,600 s (short configuration); the 43% v/v D2O contrast point was measured for 45,000 s (long) and 10,800 s (short); 80% v/v D2O, 21,600 s (long) and 10,800 s (short); 93% v/v D2O, 45,000 s (long) and 10,800 s (short) and; 100% v/v D2O: 3600 s (long) and 3600 s (short). Buffers prepared in 0% and 100% D2O were measured for 14,400 s (long) and 10,800 s (short). Supplementary files in the full entry zip archive include the buffer subtracted SANS data at each contrast point as well as SEC-SAXS data collected at SAXS-WAXS beamline (the Australian Synchrotron, ANSTO) and the subsequent results of MONSA, SASREFCV and MultiFoXS modelling. The SEC-SAXS data were collected using a sheath-flow set-up coupled to a Superdex 75 5/150 Increase column, equilibrated with 20 mM Tris, 150 mM NaCl, 2 mM TCEP, 0.1% sodium azide (pH 7.5) at a flowrate of 0.2 mL/min.
|
|
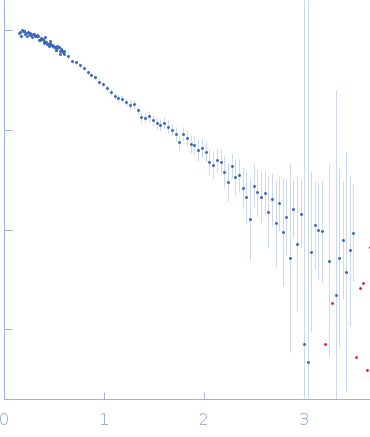 s, nm-1
s, nm-1