|
|
|
|
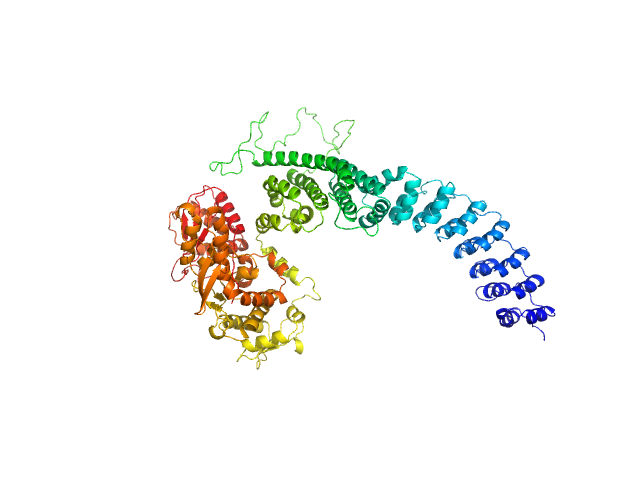
|
| Sample: |
...ubiquitin-protein ligase HACE1 monomer, 100 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 50 mM NaCl, 5 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Apr 21
|
...ubiquitin ligase HACE1
Nature Structural & Molecular Biology (2024)
Düring J, Wolter M, Toplak J, Torres C, Dybkov O, Fokkens T, Bohnsack K, Urlaub H, Steinchen W, Dienemann C, Lorenz S
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
219 |
nm3 |
|