|
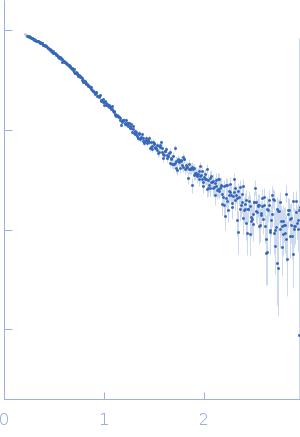
Synchrotron SAXS
data from solutions of
Diffusive GAC rRNA + Mg: Equilibrium initial
in
10 mM sodium cacodylate, 100 mM KCl, pH 6.5
were collected
on the
G1 beam line
at the Cornell High Energy Synchrotron Source (CHESS) storage ring
(Ithaca, NY, USA)
using a Pilatus 100K detector
at a wavelength of λ = 0.10972 nm
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
Solute concentrations ranging between 1.2 and 1.5 mg/ml were measured
at 20°C.
20 successive
5 second frames were collected.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
GAC rRNA (75-100 micromolar) with no added MgCl2. Molecular weight determined by Vc. Data published in Welty, R., Pabit, S. A., Katz, A. M., Calvey, G. D., Pollack, L. & Hall, K. B. (2018). RNA 24, 1828–1838. https://doi.org/10.1261/rna.068361.118
|
|
 s, nm-1
s, nm-1