|
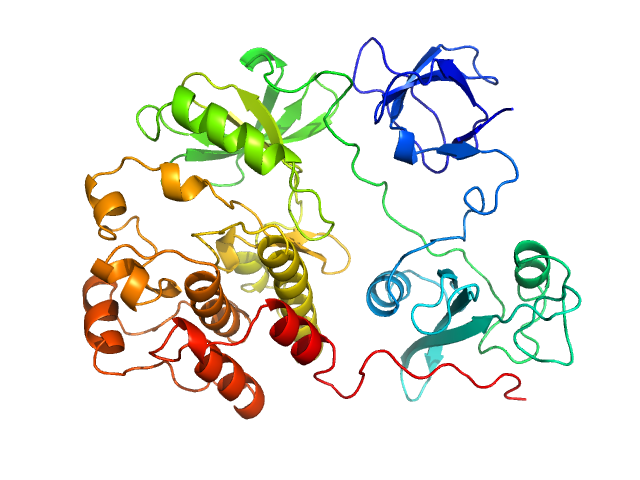
SAXS experiments were conducted using the B21 beam line at the Diamond Light Source (Didcot, UK). A sample of 40 ul of Src WT (3D-construct) at concentration of 6 mg/ml was delivered at 20 ºC via an in-line Agilent 1200 HPLC system in a Superdex 200 Increase 3.2 column (Cytiva), using a running buffer composed by 20 mM Tris pH 8.0, 150 mM NaCl, 1% glycerol and 1 mM DTT. The continuously eluting samples were exposed for 20 s and a total number of 599 frames recorded, using an X-ray wavelength of 0.1 nm, and a sample to detector (Eiger 4M) distance of 3.6 m. The frames recorded immediately before elution of the sample were subtracted from the protein scattering profiles. The Scåtter software package (www.bioisis.net) was used to analyse data, buffer-subtraction, scaling, merging and checking possible radiation damage of the samples. The Rg value was calculated with the Guinier approximation assuming that at very small angles s < 1.3/Rg. The particle distance distribution and maximum particle dimension, Dmax, were calculated from the scattering pattern with GNOM, and shape estimation was carried out with DAMMIF/DAMMIN, all these programs included in the ATSAS package. The proteins molecular mass was estimated with GNOM. A generated PDB-based homology model was made using the program COOT by manually adjusting the X-ray structure (PDB: 2SRC), into the envelope given by SAXS until a good correlation between the real-space scattering profile calculated for the homology model matched the experimental scattering data. This was computed with the program FoXS.
|
|
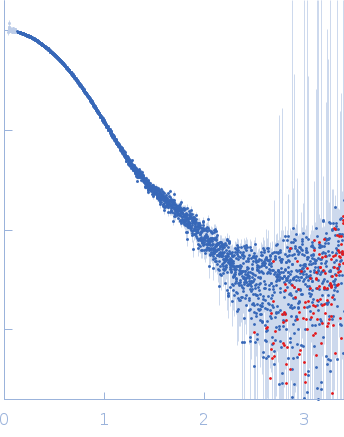 s, nm-1
s, nm-1