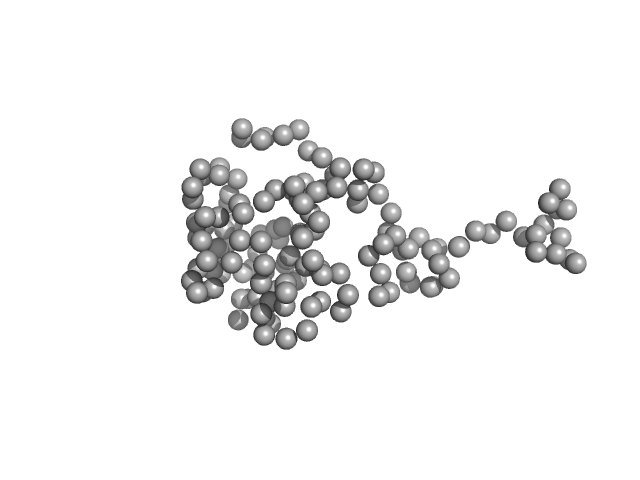|
Synchrotron SAXS
data from solutions of
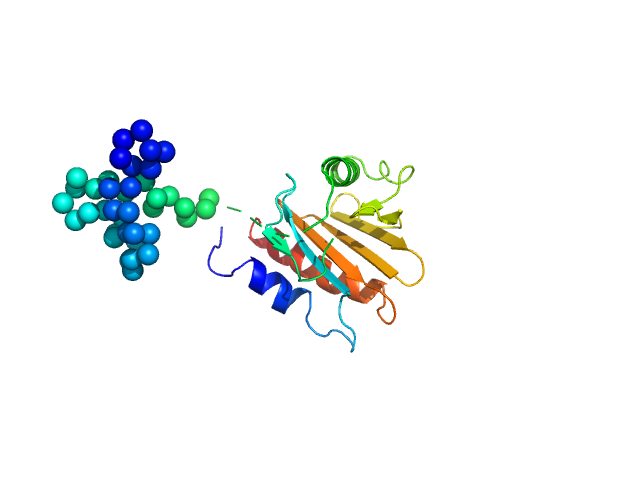
allergen Hev b 8 protein, Profilin-2 (TAG)
in
20 mM Tris, 50 mM NaCl, pH 8.4
were collected
on the
B21 beam line
at the Diamond Light Source storage ring
(Didcot, UK)
using a Eiger 4M detector
at a sample-detector distance of 3.7 m and
at a wavelength of λ = 0.094 nm
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
Solute concentrations ranging between 0.9 and 6.9 mg/ml were measured
at 20°C.
20 successive
1 second frames were collected.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
The low angle data collected at lower concentration were merged with the highest concentration high angle data to yield the final composite scattering curve.
Synchrotron SAXS data from solutions of Profilin (+TAG) in 20 mM Tris, 50 mM NaCl, pH 8.4 were collected on the B21 beam line at Diamond (Didcot, UK) using an EigerX 4M-Detrics detector at a sample-detector distance of 3.7 m and the wavelength of 0.95 A. The experiment was performed in batch at four dilution, starting at 6.9 mg/ml concentration. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted. The data didn't presented concentration dependence. Four dilution data were automatically merged by PRIMUS.
The molecular weight of a Profilin (+TAG), is 18.4 kDa. In the Ca-model (GASBOR) was possible not only to fit the PDB model (from 7sbd) but to see the TAG (40 dummy CA) as a long tail, which was next reconstructed by using of CORAL program.
|
|
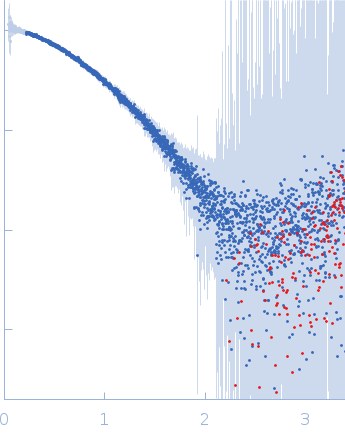 s, nm-1
s, nm-1