|
Synchrotron SAXS data from solutions of stimulatory element RNA in 20 mM HEPES pH 7.4, 400 mM KCl 2% v/v glycerol, 1 mM DTT were collected on the EMBL P12 beam line at PETRA III (DESY; Hamburg, Germany) using a Pilatus 6M detector at a sample-detector distance of 3 m and at a wavelength of λ = 0.123983 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 50.00 μl sample at 7.1 mg/ml was injected at a 0.35 ml/min flow rate onto a GE Superdex 75 Increase 5/150 column at 20°C. 57 successive 0.250 second frames were collected through the SEC-elution peak. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
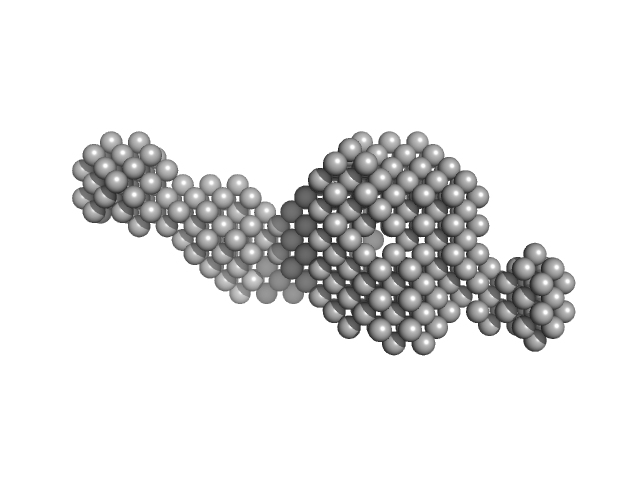
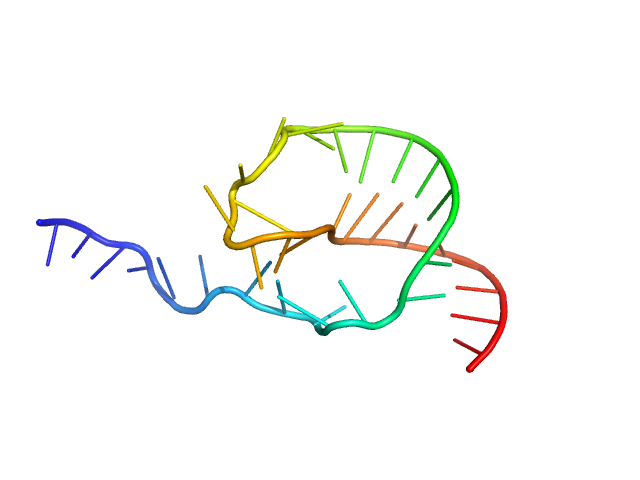
The models displayed in this entry show: Top: A post normal-mode SREFLEX model obtained using an AlphaFold-predicted structure of the RNA as an initial template, and an alignment of several different SREFLEX outputs. Middle: a MONSA ab initio dummy-atom model reconstruction of the RNA particle. Bottom: A pseudoknot conformation based on the X-ray crystal structure of the RNA in complex with Protein 2A (refer to SASBDB entry SASDWS5; https://www.sasbdb.org/data/SASDWS5). The full entry archive contains the unsubtracted SEC-SAXS 1D-data frames in addition to DAMMIN, MONSA and SREFLEX modelling routines.
|
|
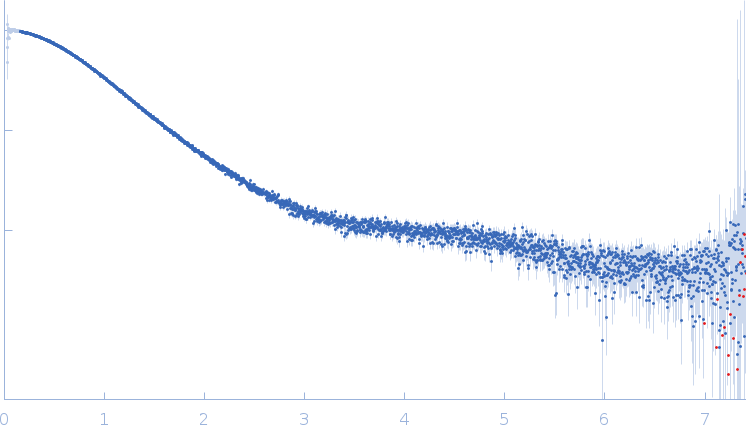 s, nm-1
s, nm-1
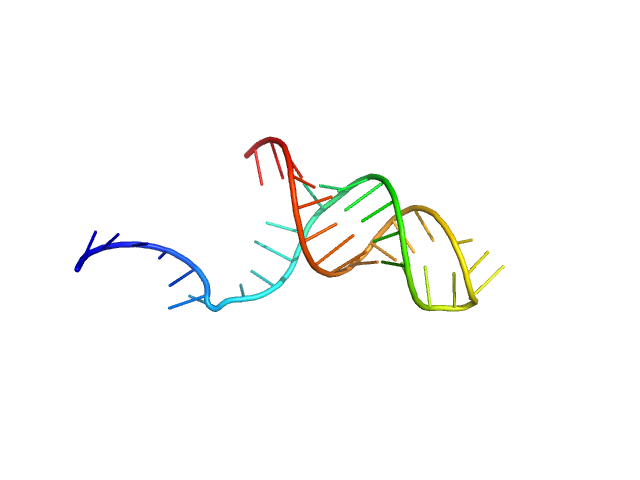
![Static model image Stimulatory element RNA OTHER [STATIC IMAGE] model](/media//pdb_file/SASDWR5_fit1_model2.png)