|
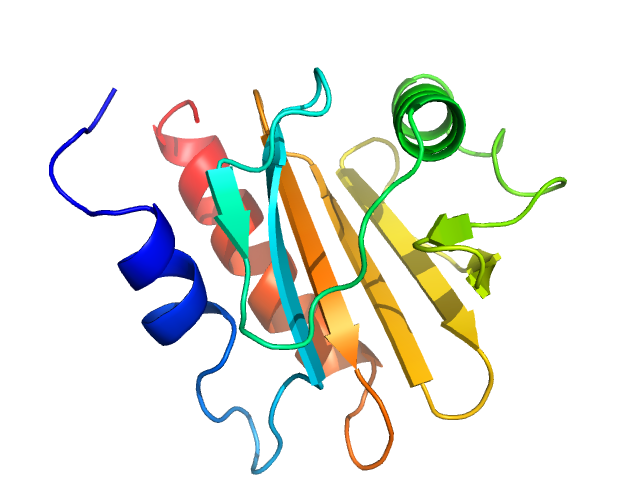
A low-resolution dummy residue model was generated by using GASBOR. In this SAXS model the crystallographic model (PDB entry 7SBD, chain C), confirms the good quality of sample preparation before running the experiments for its complex with IgE antibody.
Synchrotron SAXS data from solutions of allergen Hev b 8 protein, Profilin-2 in 20 mM Tris, 50 mM NaCl, pH 8.4 were collected on the P12 beamline at DESY (Hamburg, Germany) using a Pilatus 6m detector at a sample-detector distance of 3.0 m and a wavelength of λ = 0.124 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). A solute concentration of 0.6 mg/ml was measured at 20°C. 20 successive 0.51 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged and brought into the absolute scale with respect to the intensity of the empty capillary and the water; the scattering of the solvent-blank was subtracted.
|
|
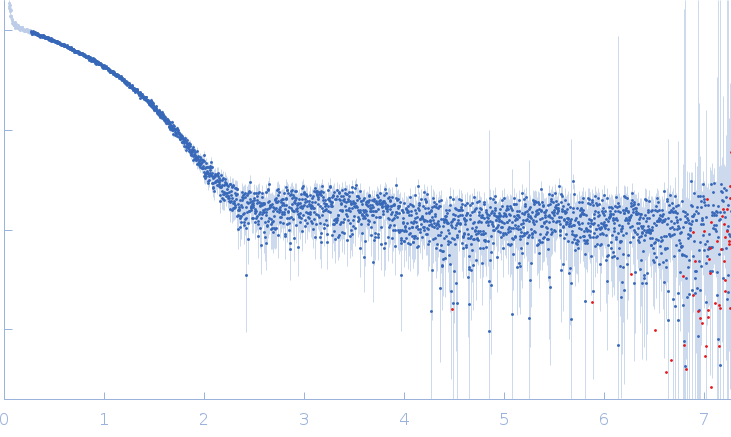 s, nm-1
s, nm-1