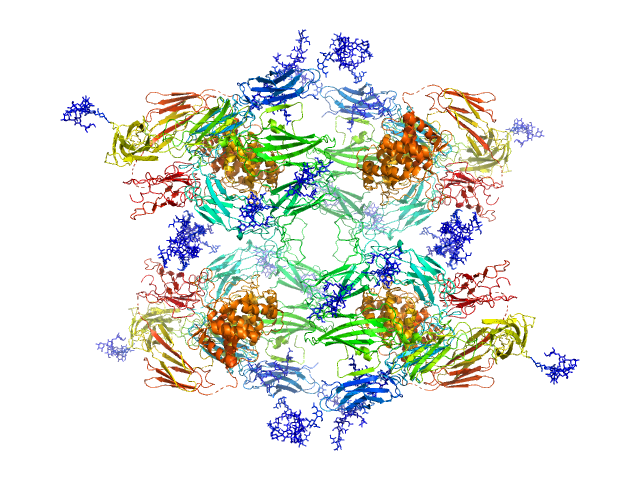
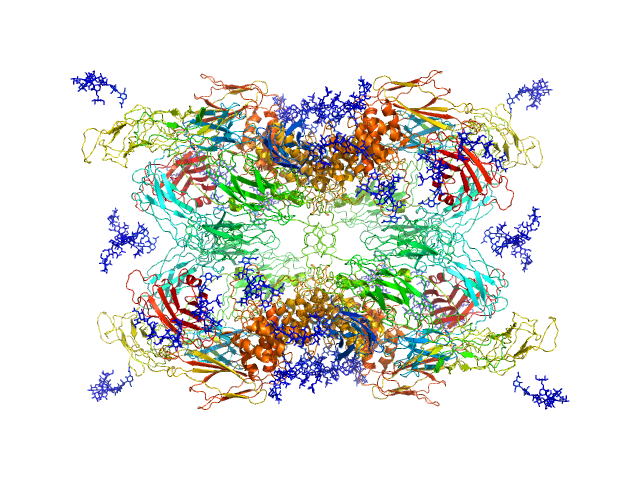
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Harwood SL,
Lyngsø J,
Zarantonello A,
Kjøge K,
Nielsen PK,
Andersen GR,
Pedersen JS,
Enghild JJ
Mol Cell Proteomics
20:100090
(2021 May 6)
|
|
|
|
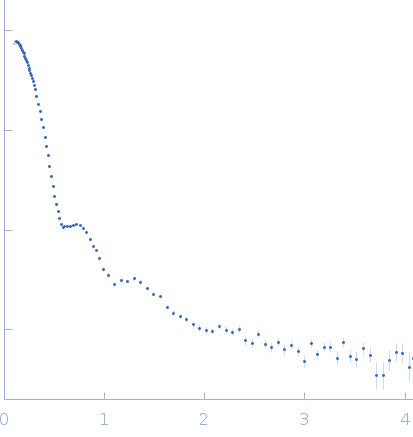
|
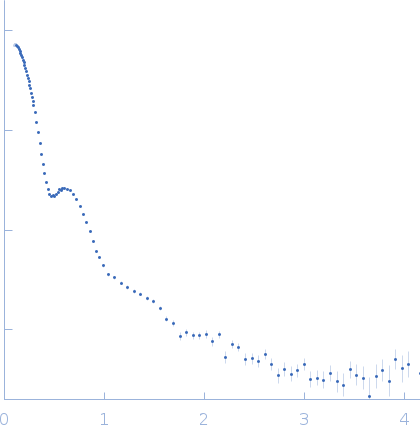
| Sample: |
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2020 Jan 22
|
|
| RgGuinier |
7.7 |
nm |
| Dmax |
22.7 |
nm |
|
|
|
|
|
|
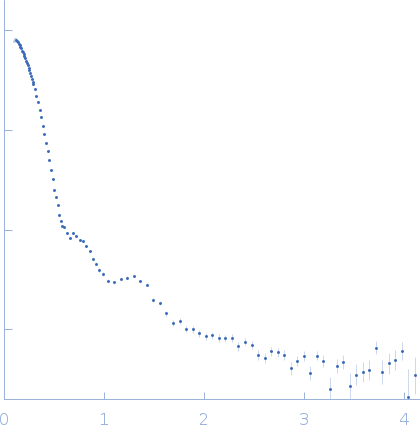
|
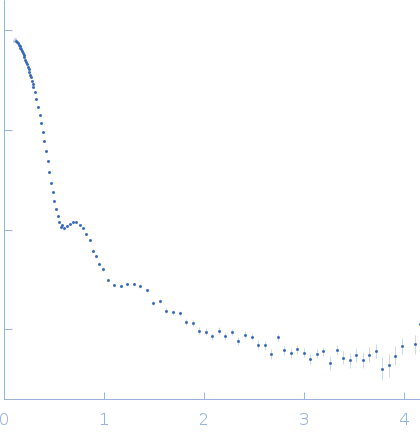
| Sample: |
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2020 Jan 22
|
|
| RgGuinier |
6.7 |
nm |
| Dmax |
19.7 |
nm |
|
|
|
|
|
|
|
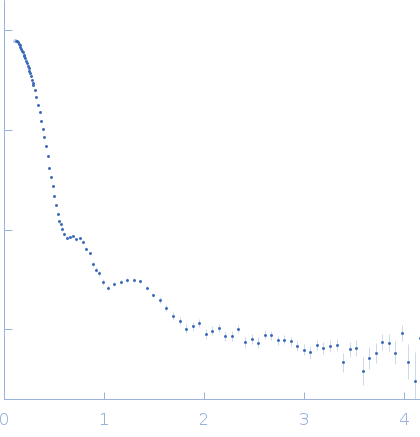
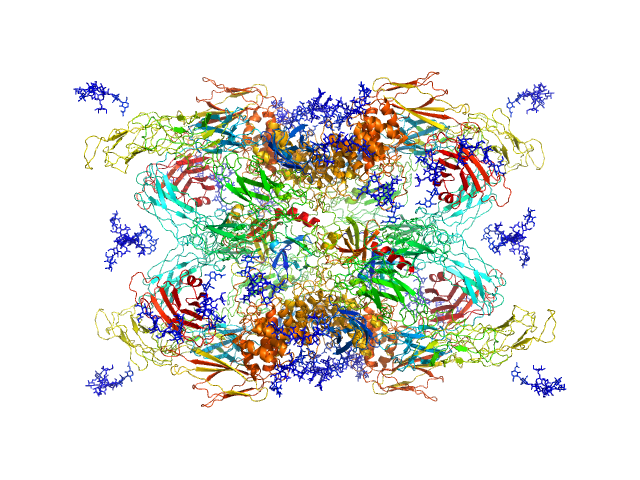
| Sample: |
Cationic trypsin dimer, 52 kDa Bos taurus protein
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2020 Jan 15
|
|
| RgGuinier |
6.6 |
nm |
| Dmax |
19.7 |
nm |
|
|
|
|
|
|
|
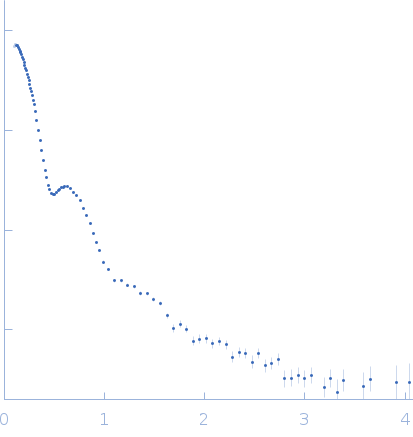
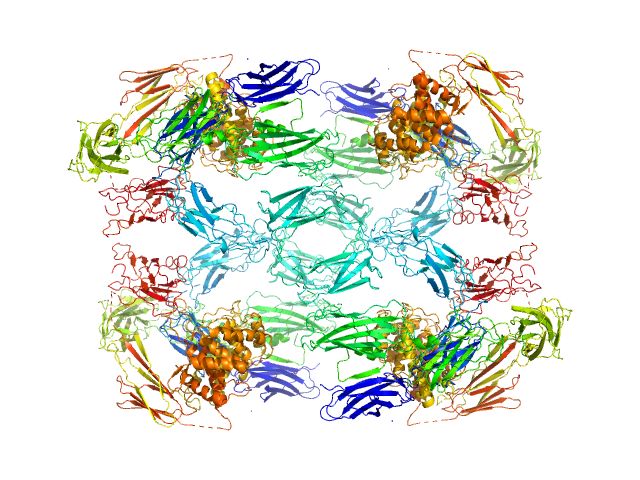
| Sample: |
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2019 Apr 12
|
|
| RgGuinier |
7.6 |
nm |
| Dmax |
20.2 |
nm |
|
|
|
|
|
|
|
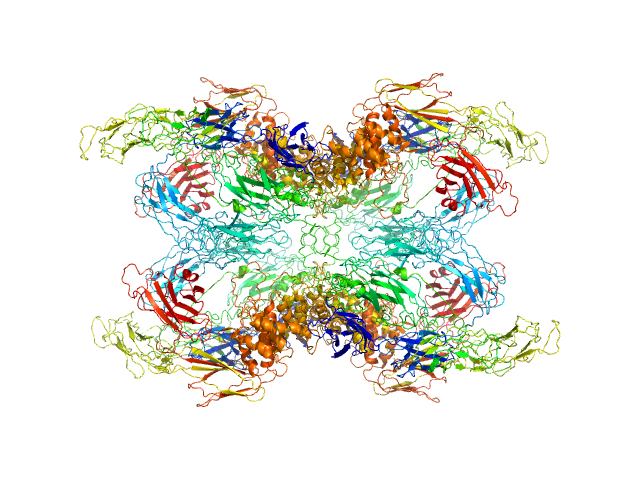
| Sample: |
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2019 Apr 13
|
|
| RgGuinier |
6.6 |
nm |
| Dmax |
19.7 |
nm |
|
|
|
|
|
|
|
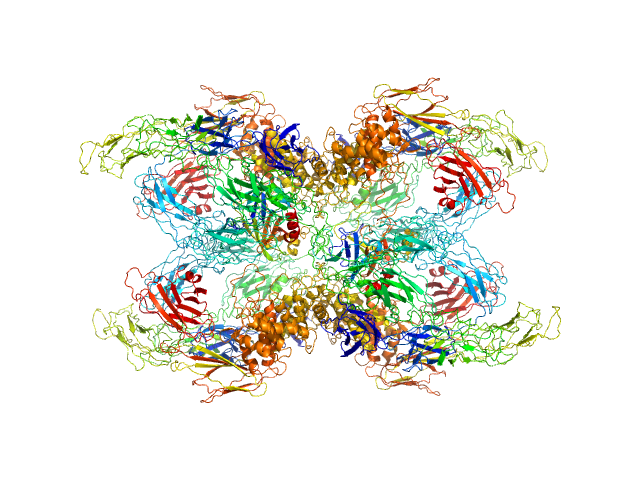
| Sample: |
Cationic trypsin dimer, 52 kDa Bos taurus protein
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2020 Jan 22
|
|
| RgGuinier |
6.6 |
nm |
| Dmax |
20.7 |
nm |
|
|