Structural basis for the role of C-terminus acidic tail of Saccharomyces cerevisiae ubiquitin-conjugating enzyme (Rad6) in E3 ligase (Bre1) mediated recognition of histones
Yadav P,
Gupta M,
Wazahat R,
Islam Z,
Tsutakawa S,
Kamthan M,
Kumar P
International Journal of Biological Macromolecules
:127717
(2023 Nov)
|
|
|
|
|
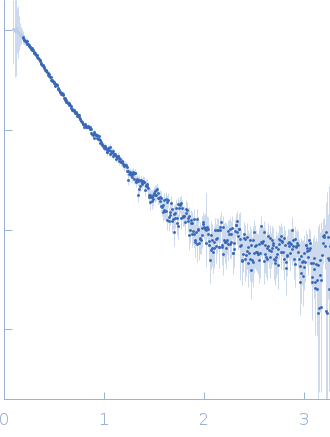
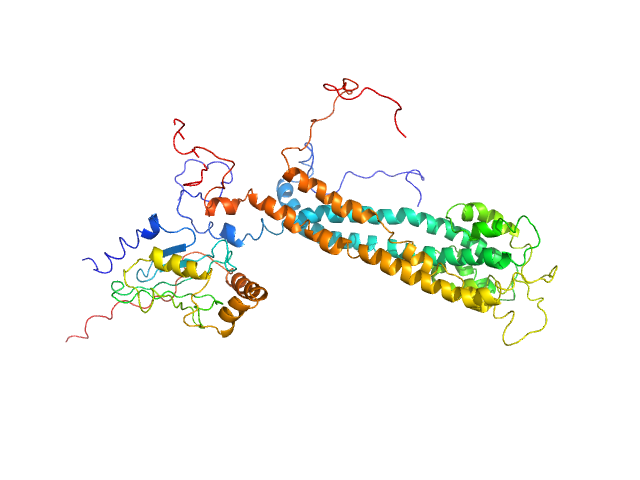
| Sample: |
E3 ubiquitin-protein ligase BRE1 dimer, 50 kDa Saccharomyces cerevisiae (strain … protein
Ubiquitin-conjugating enzyme E2 2 monomer, 20 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2014 Feb 11
|
|
| RgGuinier |
4.1 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
|
|
|
|
|
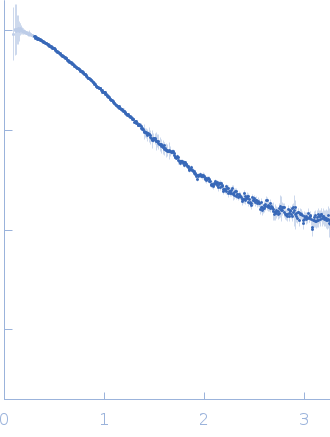
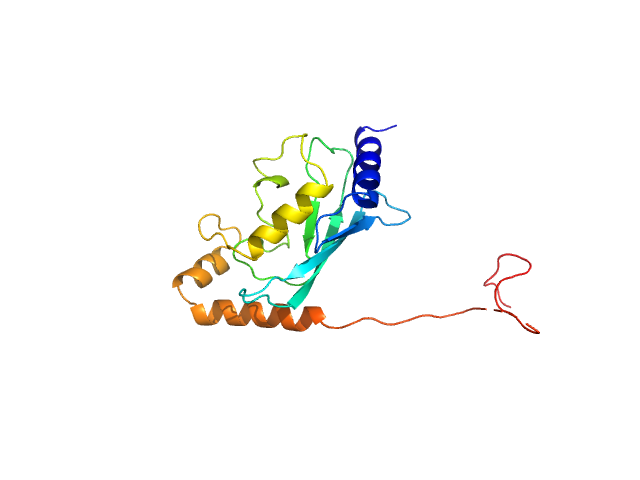
| Sample: |
Ubiquitin-conjugating enzyme E2 2 monomer, 20 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2014 Feb 11
|
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
31 |
nm3 |
|
|