Protein quaternary structures in solution are a mixture of multiple forms
Marciano S,
Dey D,
Listov D,
Fleishman S,
Sonn-Segev A,
Mertens H,
Busch F,
Kim Y,
Harvey S,
Wysocki V,
Schreiber G
Chemical Science
13(39):11680-11695
(2022)
|
|
|
|
|
| Sample: |
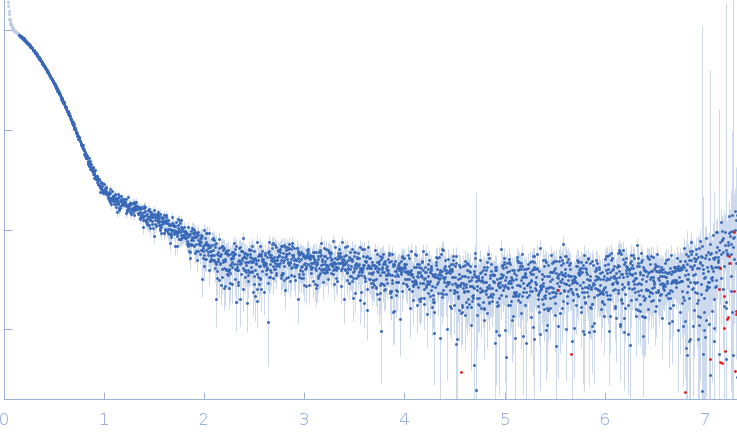
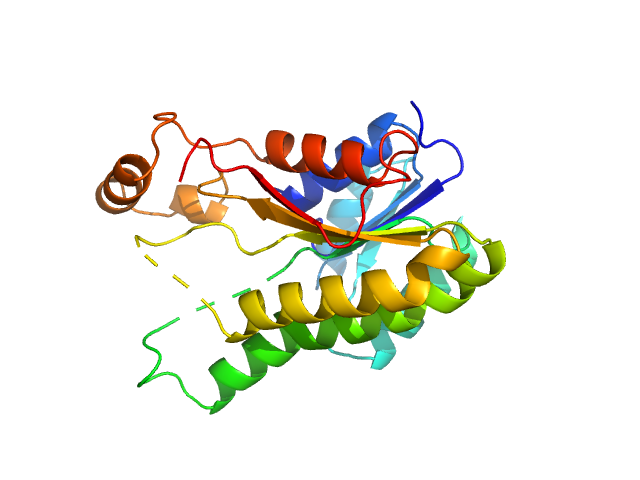
3-oxoacyl-[acyl-carrier-protein] reductase FabG, 26 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Oct 1
|
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
212 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
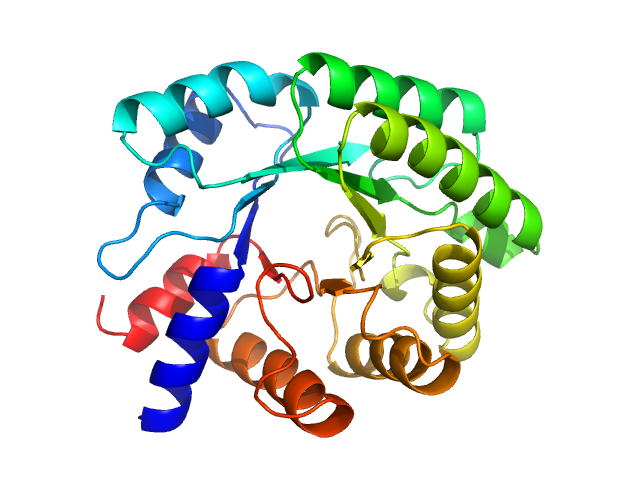
Deoxyribose-phosphate aldolase, 28 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Oct 1
|
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
|
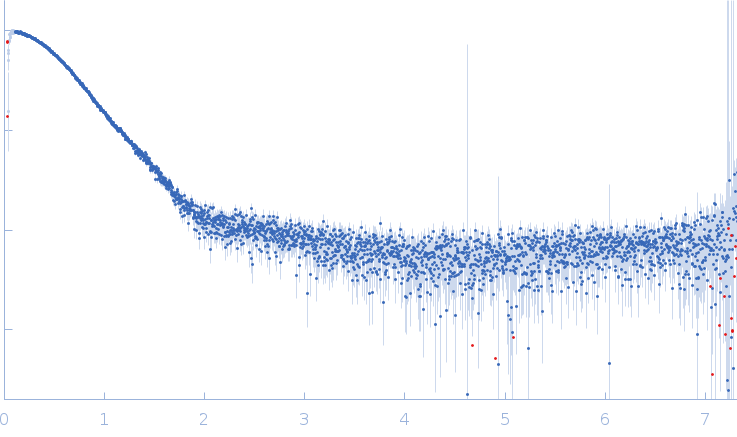
| Sample: |
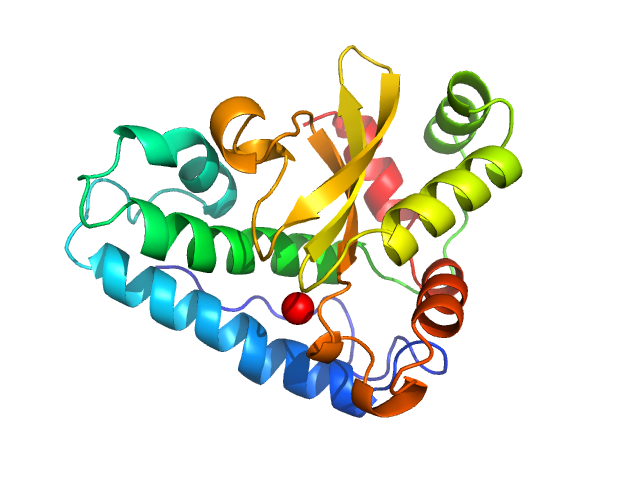
Superoxide dismutase [Mn], 23 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Oct 1
|
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
|
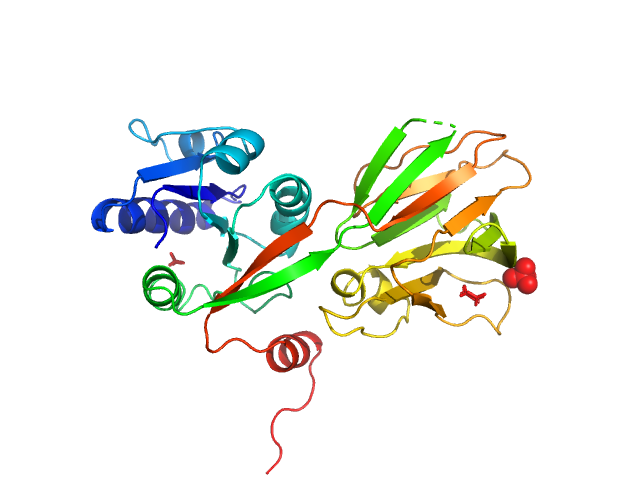
| Sample: |
NAD kinase, 33 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM HEPES, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Oct 2
|
|
| RgGuinier |
4.0 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
260 |
nm3 |
|
|

![3-oxoacyl-[acyl-carrier-protein] reductase FabG experimental SAS data 3-oxoacyl-[acyl-carrier-protein] reductase FabG experimental SAS data](/media/intensities_files/scattering_plots/SASDLR4_dat_img.png)


![Superoxide dismutase [Mn] experimental SAS data Superoxide dismutase [Mn] experimental SAS data](/media/intensities_files/scattering_plots/SASDLP4_dat_img.png)