Structural insights into the substrate binding mechanism of the class I dehydratase MadB
Knospe C,
Ortiz J,
Reiners J,
Kedrov A,
Gertzen C,
Smits S,
Schmitt L
Communications Biology
8(1)
(2025 Jul 09)
|
|
|
|
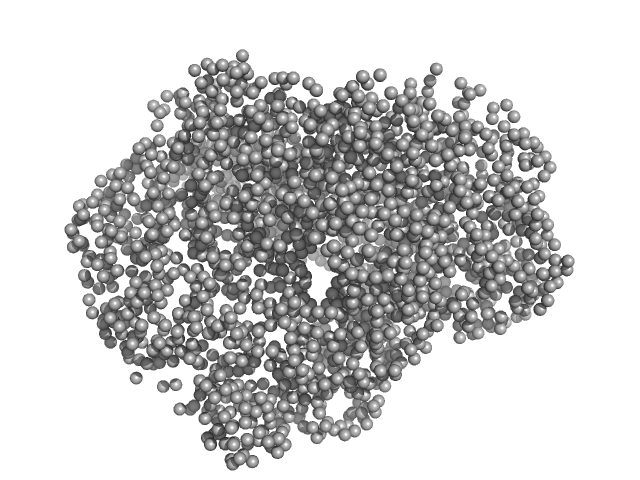
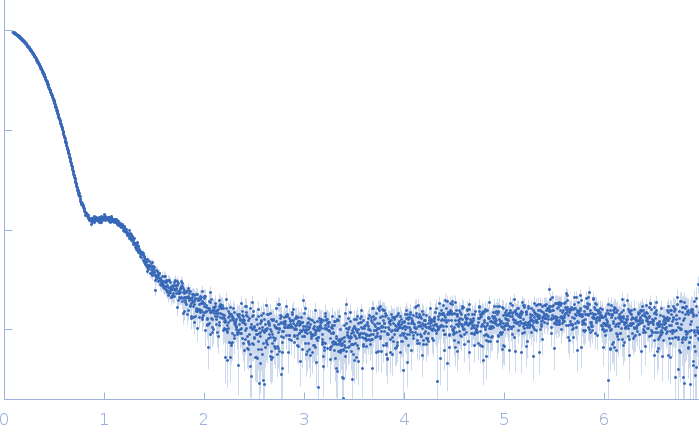
|
| Sample: |
Thiopeptide-type bacteriocin biosynthesis domain containing protein dimer, 250 kDa Clostridium sp. Maddingley … protein
|
| Buffer: |
50 mM HEPES, 200 mM NaCl, 1 % (v/v) Glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 26
|
|
| RgGuinier |
4.5 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
440 |
nm3 |
|
|
|
|
|
|
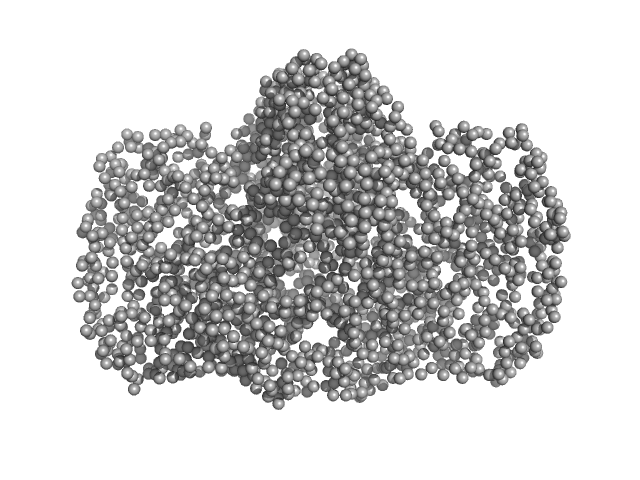
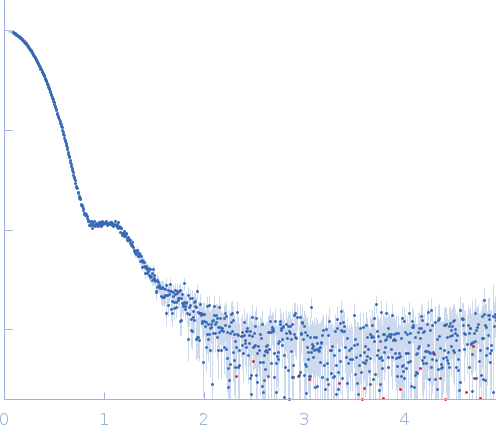
|
| Sample: |
Thiopeptide-type bacteriocin biosynthesis domain containing protein dimer, 250 kDa Clostridium sp. Maddingley … protein
Lantibiotic, gallidermin/nisin family monomer, 3 kDa Clostridium sp. Maddingley … protein
|
| Buffer: |
50 mM HEPES, 200 mM NaCl, 1 % (v/v) Glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 3
|
|
| RgGuinier |
4.5 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
445 |
nm3 |
|
|