|
|
|
|
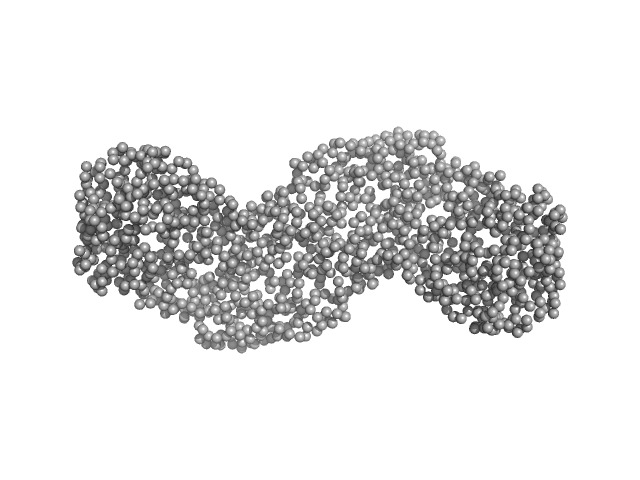
|
| Sample: |
Gbp6 protein (Gbp7 - Gbp6 protein Mus musculus) dimer, 148 kDa Mus musculus protein
|
| Buffer: |
50 mM Tris, 5 mM MgCl, 2 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2019 Oct 2
|
Integrative modeling of guanylate binding protein dimers
Protein Science (2023)
...Reiners J, Degrandi D, Legewie L, Stühler K, Pfeffer K, Poschmann G, Smits S, Strodel B
|
| RgGuinier |
5.4 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
176 |
nm3 |
|