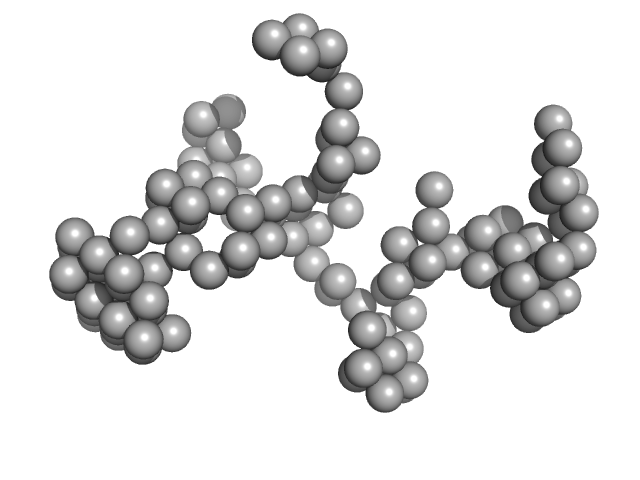Structural Dynamics of Dengue Virus UTRs and Their Cyclization.
Robinson ZE,
Pereira HS,
D'Souza MH,
Patel TR
Biophys J
(2025 Sep 5)
|
|
|
|
|
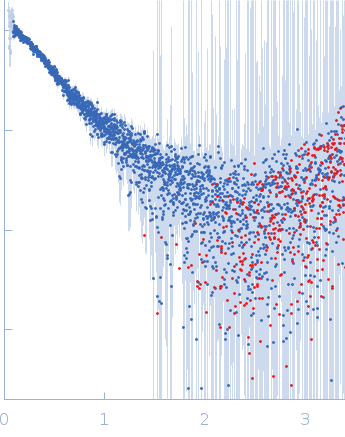
| Sample: |
DENV2 RNA, 5' Untranslated region monomer, 57 kDa RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, 5 mM MgCl2, 15 mM KCl, 5% glycerol, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 8
|
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
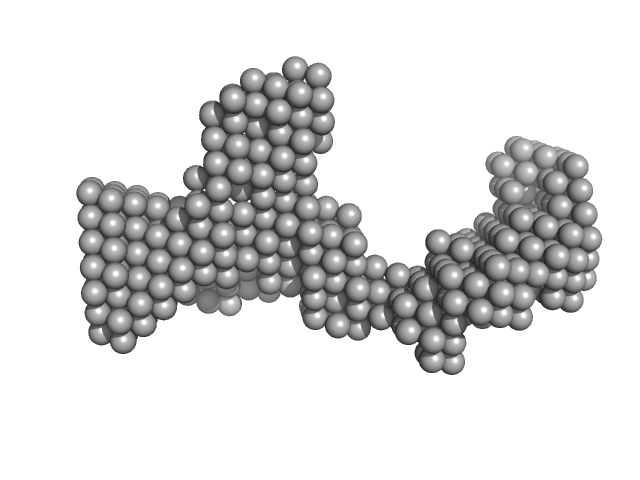
|
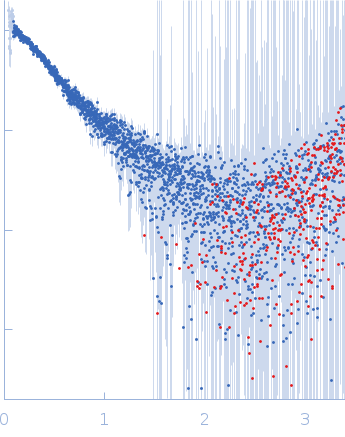
| Sample: |
DENV2 5' untranslated terminal region monomer, 57 kDa RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, 5 mM MgCl2, 15 mM KCl, 5% glycerol, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 10
|
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|
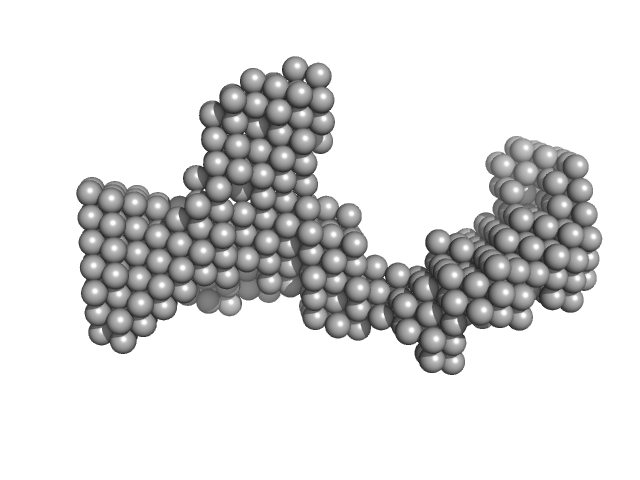
|
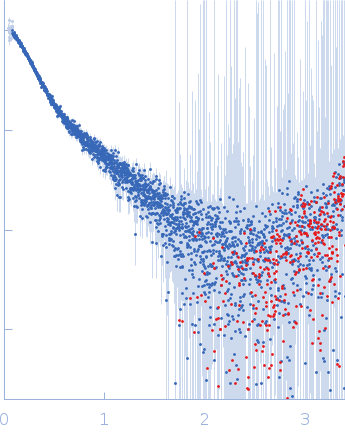
| Sample: |
DENV2 3' untranslated region monomer, 160 kDa RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, 5 mM MgCl2, 15 mM KCl, 5% glycerol, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 10
|
|
| RgGuinier |
6.0 |
nm |
| Dmax |
18.7 |
nm |
| VolumePorod |
155 |
nm3 |
|
|
|
|
|
|
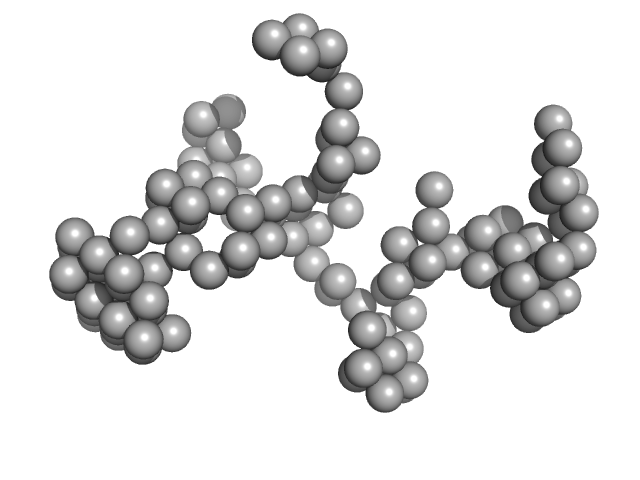
|
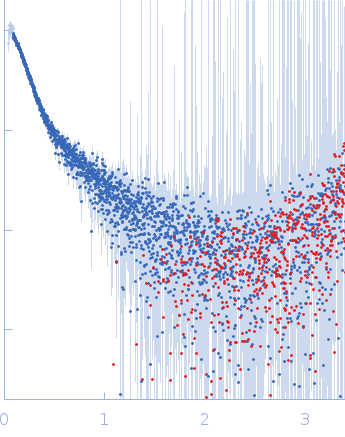
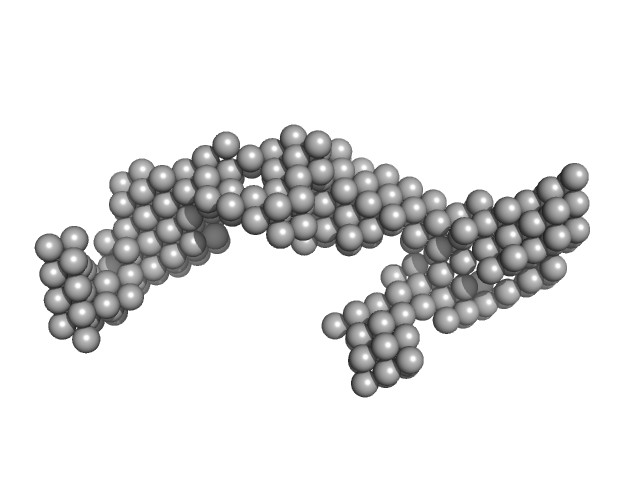
| Sample: |
DENV2 complex 5'-3' monomer, 217 kDa RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, 5 mM MgCl2, 15 mM KCl, 5% glycerol, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 10
|
|
| RgGuinier |
7.5 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
320 |
nm3 |
|
|