|
|
|
|
|
| Sample: |
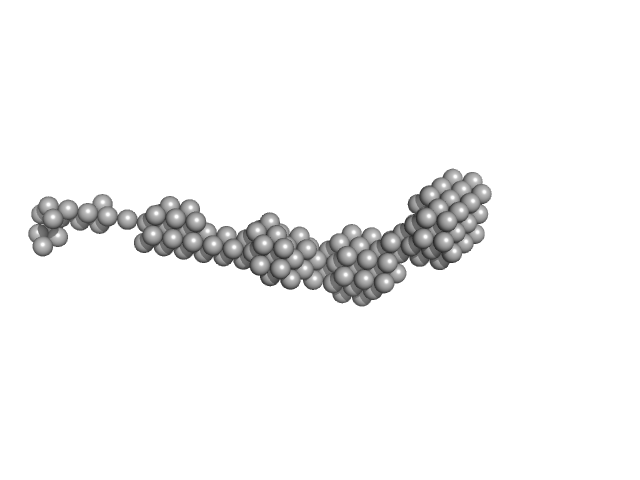
Bromodomain-containing protein 4 monomer, 56 kDa Homo sapiens protein
|
| Buffer: |
20mM Hepes, 100mM NaCl, 1mM Tris(2-carboxyethyl)phosphine hydrochloride, pH: 7.4
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 12
|
Potent and selective bivalent inhibitors of BET bromodomains.
Nat Chem Biol 12(12):1097-1104 (2016)
...Patel J, Petteruti P, Robb GR, Robers MB, Saif S, Stratton N, Svergun DI, Wang W, Whittaker D, Wilson DM, Yao Y
|
| RgGuinier |
6.2 |
nm |
| Dmax |
27.5 |
nm |
|