Molecular basis of Fab-dependent IgA antibody recognition by gut-bacterial metallopeptidases
Márquez-Moñino M,
Martínez Gascueña A,
Azzam T,
Persson A,
Manzanares-Gomez A,
Aguillo-Urarte M,
Brown T,
Montero-Sagarminaga A,
Lood R,
Naegeli A,
Connell S,
Sastre D,
Sundberg E,
Trastoy B
The EMBO Journal
(2025 Jul 31)
|
|
|
|
|
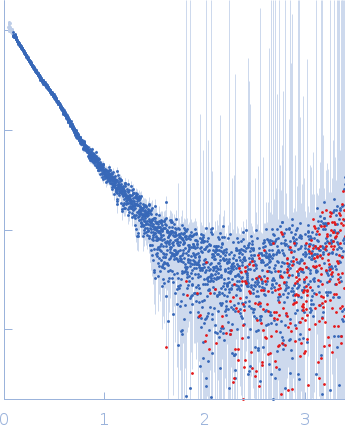
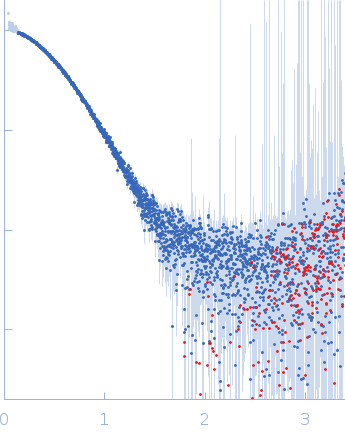
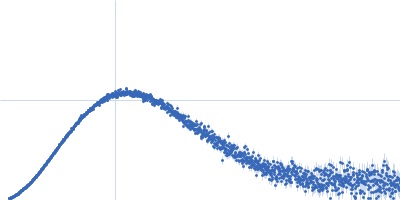
| Sample: |
IgA protease monomer, 127 kDa Thomasclavelia ramosa protein
|
| Buffer: |
50 mM Tris-HCl pH 7.5, 100 mM NaCl, 2% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Apr 19
|
|
| RgGuinier |
6.4 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
208 |
nm3 |
|
|
|
|
|
|
|
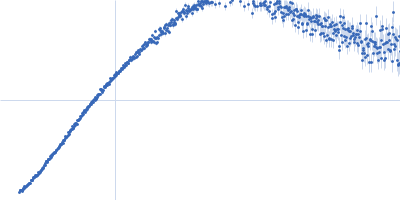
| Sample: |
IgA protease monomer, 96 kDa Thomasclavelia ramosa protein
|
| Buffer: |
50 mM Tris-HCl pH 7.5, 100 mM NaCl, 2% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Apr 19
|
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
101 |
nm3 |
|
|
|
|
|
|
|
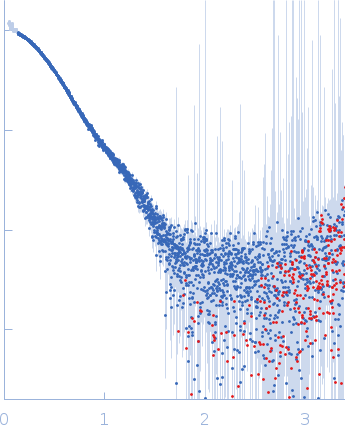
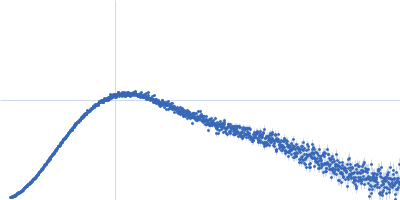
| Sample: |
IgA protease monomer, 64 kDa Thomasclavelia ramosa protein
|
| Buffer: |
50 mM Tris-HCl pH 7.5, 100 mM NaCl, 2% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Apr 19
|
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
72 |
nm3 |
|
|