Analysis of the natively unstructured RNA/protein-recognition core in the Escherichia coli RNA degradosome and its interactions with regulatory RNA/Hfq complexes.
Bruce HA,
Du D,
Matak-Vinkovic D,
Bandyra KJ,
Broadhurst RW,
Martin E,
Sobott F,
Shkumatov AV,
Luisi BF
Nucleic Acids Res
46(1):387-402
(2018 Jan 9)
|
|
|
|
|
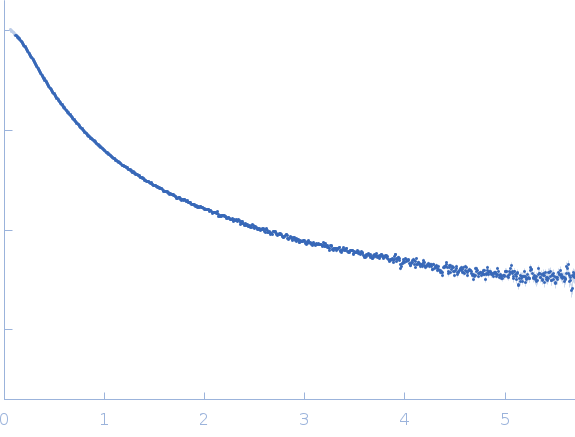
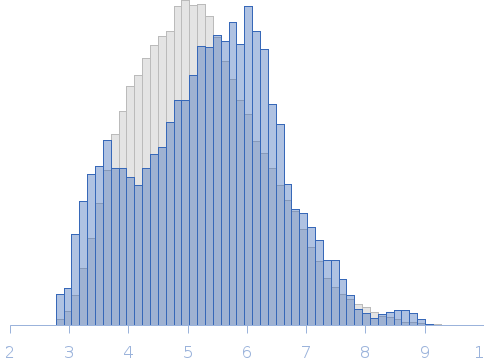
| Sample: |
RNase E 603-850 monomer, 30 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris HCl, 100 mM NaCl, 100 mM KCl, 10 mM MgCl2, 10 mM DTT and 5 % glycerol (v/v), pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Dec 5
|
|
| RgGuinier |
5.3 |
nm |
| Dmax |
27.5 |
nm |
| VolumePorod |
139 |
nm3 |
|
|
|
|
|
|

|
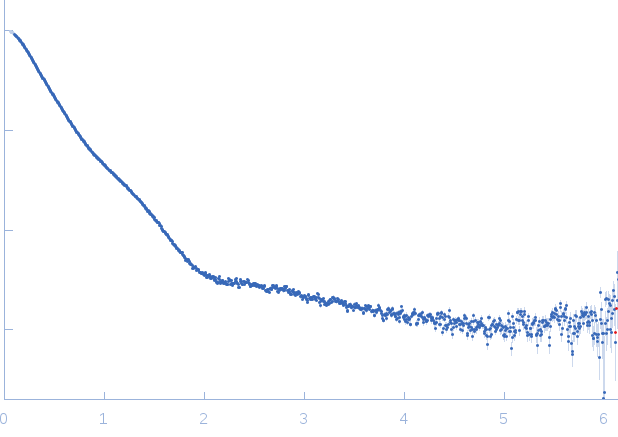
| Sample: |
RNase E 603-850 monomer, 30 kDa Escherichia coli protein
ATP-dependent RNA helicase RhlB monomer, 47 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris HCl, 100 mM NaCl, 100 mM KCl, 10 mM MgCl2, 10 mM DTT and 5 % glycerol (v/v), pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Feb 11
|
|
| RgGuinier |
5.4 |
nm |
| Dmax |
29.5 |
nm |
| VolumePorod |
183 |
nm3 |
|
|
|
|
|
|

|
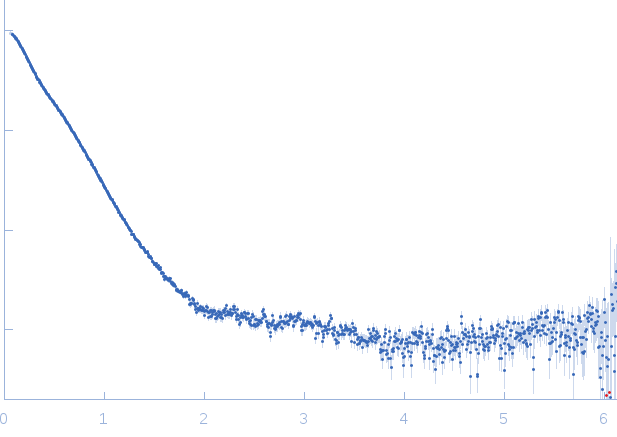
| Sample: |
RNase E 603-850 monomer, 30 kDa Escherichia coli protein
ATP-dependent RNA helicase RhlB monomer, 47 kDa Escherichia coli protein
Enolase dimer, 91 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris HCl, 100 mM NaCl, 100 mM KCl, 10 mM MgCl2, 10 mM DTT and 5 % glycerol (v/v), pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Jul 16
|
|
| RgGuinier |
6.4 |
nm |
| Dmax |
30.5 |
nm |
| VolumePorod |
280 |
nm3 |
|
|