|
|
|
|
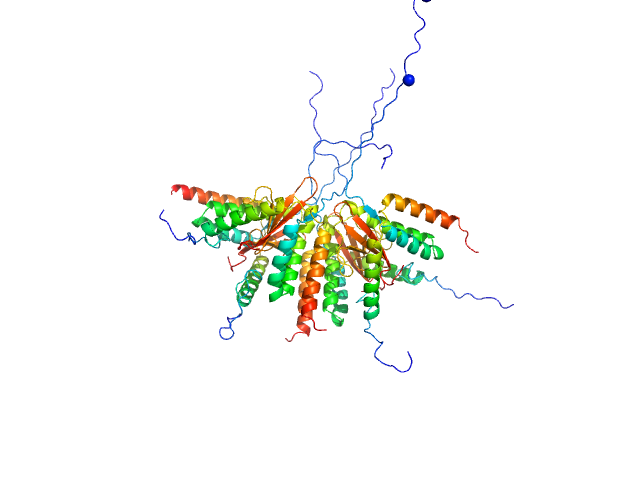
|
| Sample: |
Plasmid stabilization protein ParE octamer, 102 kDa Escherichia coli protein
Uncharacterized protein (Antitoxin) octamer, 68 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris-HCl 500 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 Feb 5
|
A unique hetero-hexadecameric architecture displayed by the Escherichia coli O157 PaaA2-ParE2 antitoxin-toxin complex.
J Mol Biol 428(8):1589-603 (2016)
...Shkumatov AV, Garcia-Pino A, Geerts L, De Kerpel M, Lah J, De Greve H, Van Melderen L, Loris R
|
| RgGuinier |
3.3 |
nm |
| Dmax |
15.3 |
nm |
| VolumePorod |
166 |
nm3 |
|