|
|
|
|
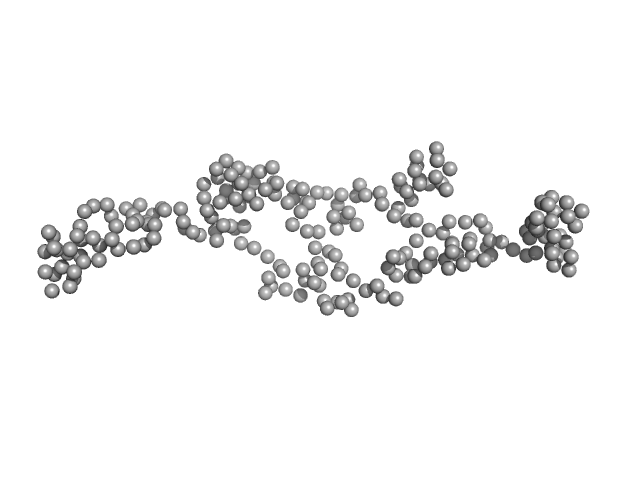
|
| Sample: |
AT5g04600/T32M21_200 monomer, 25 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM HNa2PO4, 300 mM NaCl, 5% glycerol (v/v), 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 22
|
Structural and functional analysis of a plant nucleolar RNA chaperone-like protein.
Sci Rep 13(1):9656 (2023)
Fernandes R, Ostendorp A, Ostendorp S, Mehrmann J, Falke S, Graewert MA, Weingartner M, Kehr J, Hoth S
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
66 |
nm3 |
|