|
|
|
|
|
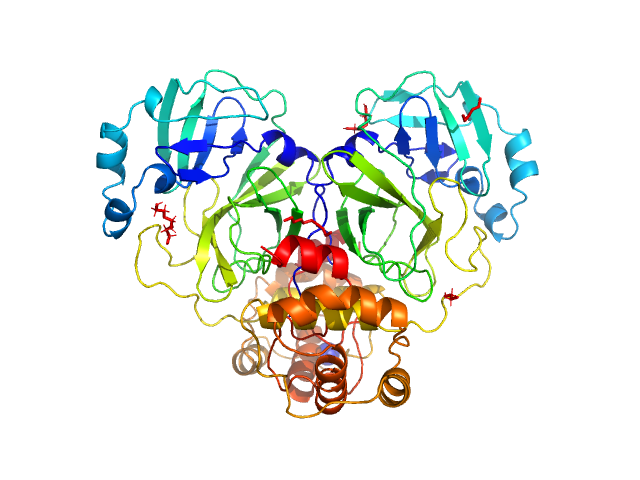
| Sample: |
Replicase polyprotein 1ab, H3426A (3C-like proteinase nsp5 - H163A mutant) dimer, 67 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM HEPES, 1 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Mar 24
|
The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun 14(1):5625 (2023)
Tran N, Dasari S, Barwell SAE, McLeod MJ, Kalyaanamoorthy S, Holyoak T, Ganesan A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
102 |
nm3 |
|