|
|
|
|
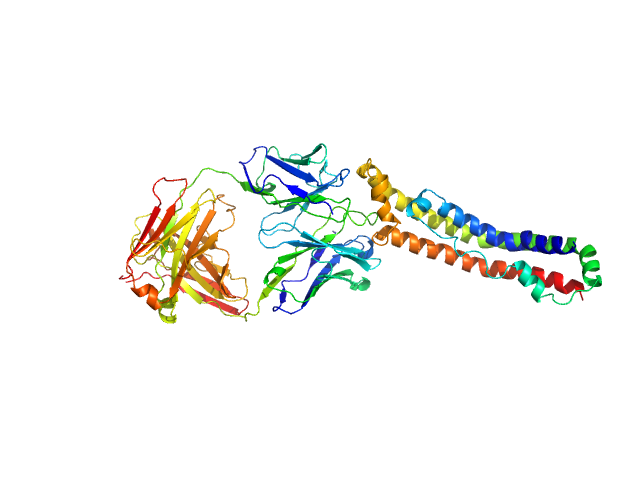
|
| Sample: |
Apolipoprotein A-I monomer, 28 kDa Homo sapiens protein
Antigen-binding fragment 55201 monomer, 45 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.1% sodium azide, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Jun 19
|
...apolipoprotein A-I monomer provides insights into its oligomerisation and lipid-binding mechanisms
Journal of Molecular Biology :169394 (2025)
Tou H, Rosenes Z, Khandokar Y, Zlatic C, Metcalfe R, Mok Y, Morton C, Gooley P, Griffin M
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
99 |
nm3 |
|