|
|
|
|
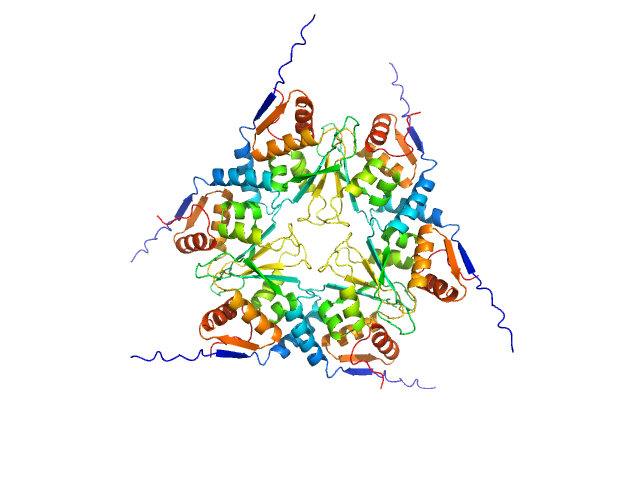
|
| Sample: |
Uncharacterized protein, isoform A hexamer, 92 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jul 20
|
The Arthropoda-specific Tramtrack group BTB protein domains use previously unknown interface to form hexamers.
Elife 13 (2024)
Bonchuk AN, Balagurov KI, Baradaran R, Boyko KM, Sluchanko NN, Khrustaleva AM, Burtseva AD, Arkova OV, Khalisova KK, Popov VO, Naschberger A, Georgiev PG
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
166 |
nm3 |
|