|
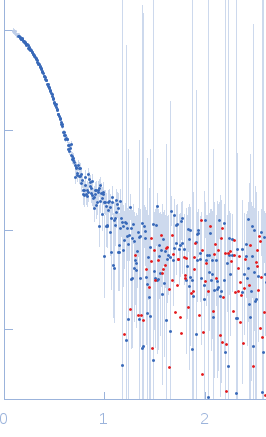
X-ray synchrotron radiation scattering data from a solution of E. coli TraE protein in 50 mM sodium phosphate, 300 mM NaCl, 40 mM imidazole, 0.15 % octyl glucose neopentyl glycol (OGNG), pH 7.4 were collected using size-exclusion chromatography (SEC) SAXS on the G1 beam line at the CHESS storage ring (Ithaca, USA) using a CCD Finger Lakes CCD detector (I(s) vs s, where s = 4π sin θ/λ; 2θ is the scattering angle; λ = 0.12 nm). The data were collected as 70 successive 2 second frames through the hexameric TraE protein/OGNG elution peak and were normalized to the intensity of the transmitted beam and radially averaged. The scattering of the solvent-blank was subtracted to produce the scattering profile displayed in this entry.
The MW of the TraE/OGNG complex was additionally confirmed using SEC-MALLS (248kDa).
|
|
 s, nm-1
s, nm-1
