|
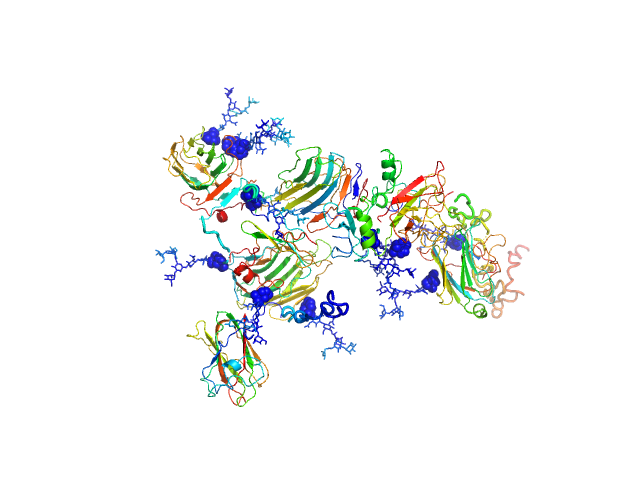
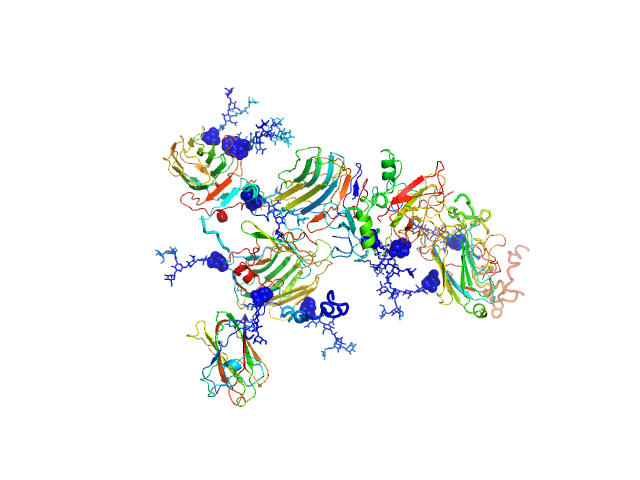
SAXS data were collected from solutions of contactin-associated protein-like 2 (Caspr2, amino acids 1-1261) in 10 mM HEPES 150 mM NaCl using an Anton Paar SAXSess line collimation instrument (University of Utah, USA). Scattering intensities, I(s) vs s (where s = 4π sin θ/λ, 2θ is the scattering angle, λ = 0.154 nm, CuKα) were recorded on image plates using a 10 mm slit geometry for 30 minutes (20°C sample temperature). A 10 mm integration width was employed to produce averaged 1D-smeared data that were corrected for beam intensity and beam geometry to generate the 1D-desmeared SAXS data for this entry. Different solute concentrations were measured in the range of 1.9-9.6 mg/ml. The data, P(r) vs r, ab initio bead model and rigid-body models shown above report the results obtained for the 7.7 mg/ml sample ('0.80' dilution.) All additional data, including the desmeared data for the diltution series (1, 0.8, 0.6, 0.4 and 0.2), the P(r) vs r calculated using GNOM and GIFT as well as all DAMMIN, DAMMIF (ab initio) and CORAL rigid-body models are included in the full-entry.zip archive. Both CRYSOL and CORAL fits to the data for the rigid-body models are included. Note: although one Caspr2 model is presented above, the protein likely samples several conformational states in solution as evidenced by the variation in the models generated during the course of ab initio and rigid-body modelling (refer to the full-entry archive).
|
|
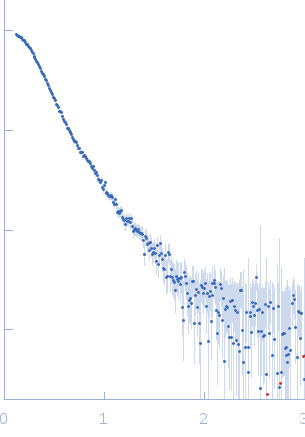 s, nm-1
s, nm-1