|
|
|
|
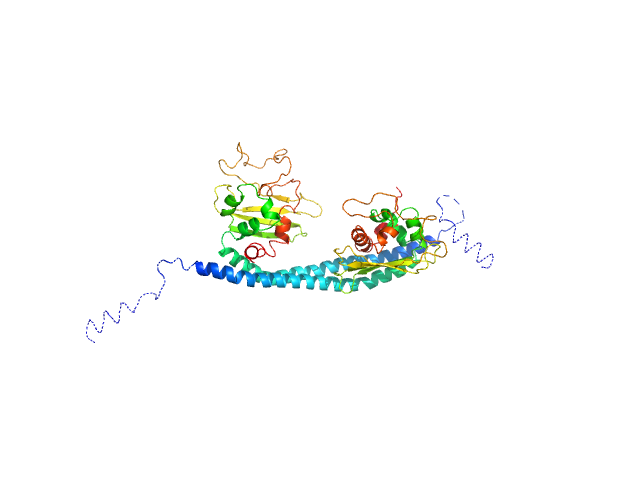
|
| Sample: |
SET nuclear proto-oncogene dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
Sodium phosphate buffer, pH: 6.3
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 11
|
PP2A is activated by cytochrome c upon formation of a diffuse encounter complex with SET/TAF-Iβ
Computational and Structural Biotechnology Journal (2022)
...Molodenskiy D, Díaz-Quintana A, Martinho M, Gerbaud G, González-Arzola K, Velázquez-Campoy A, Svergun D, Belle V, De la Rosa M, Díaz-Moreno I
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
87 |
nm3 |
|