|
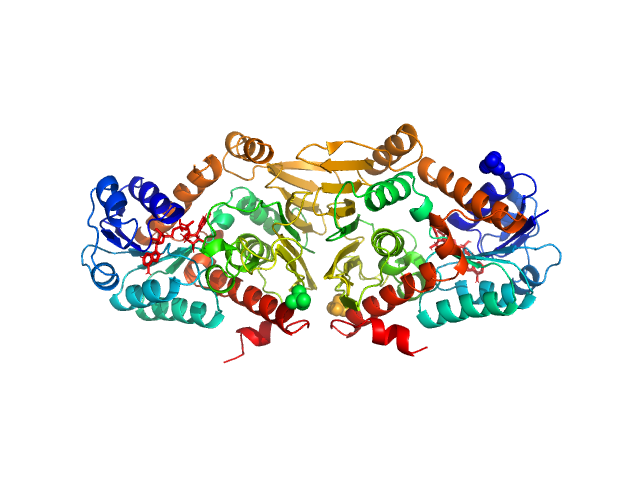
Synchrotron SAXS data of the oxidoreductase YjhC from Escherichia coli in 20 mM Tris, 150 mM NaCl, 0.1 % (w/v) sodium azide, 5 % (v/v) glycerol, pH 8 were collected on the SAXS/WAXS beam line at the Australian Synchrotron (Melbourne, Australia) using a PILATUS 1M detector at a sample-detector distance of 1.6 m, providing a q range of 0.06-5 nm-1 and at a wavelength of λ = 0.10332 nm (l(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). Coflow size-exclusion chromatography-SAXS (SEC-SAXS) was used to minimize sample dilution and maximize signal to noise (Kirby et al., 2016). 80 µl of purified YjhC protein at 8 mg/ml was injected onto an Superdex S200 Increase 5/150 GL (GE Healthcare) at a flow rate of 0.45 ml/min. Scattering data was collected in 1 second exposures over a total of 400 frames, using a 1.5 mm glass capillary, at 12 °C. 2D intensity plots were radially averaged, normalised to sample transmission, and background subtracted using the Scatterbrain software package (Australian Synchrotron). Analysis of the scattering data was performed using the ATSAS software package (version 2.8.4). PrimusQT was used to perform the Guinier analysis, Kratky analysis, and to generate the pairwise distribution function P(r), calculated using an indirect Fourier transform using GNOM. The molecular mass of the samples was estimated using the SAXS-MoW2 package (Fischer et al., 2010), while CRYSOL was used to evaluate the experimental solution scattering with the calculated scattering from the atomic coordinates of the high-resolution structure of YjhC.
SEC column = UNKNOWN. Sample injection volume = UNKNOWN. Flow rate = UNKNOWN
|
|
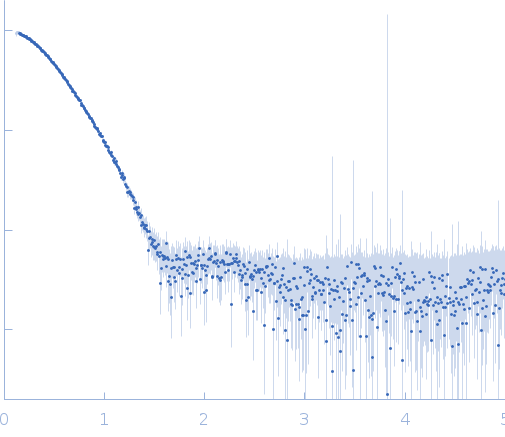 s, nm-1
s, nm-1