|
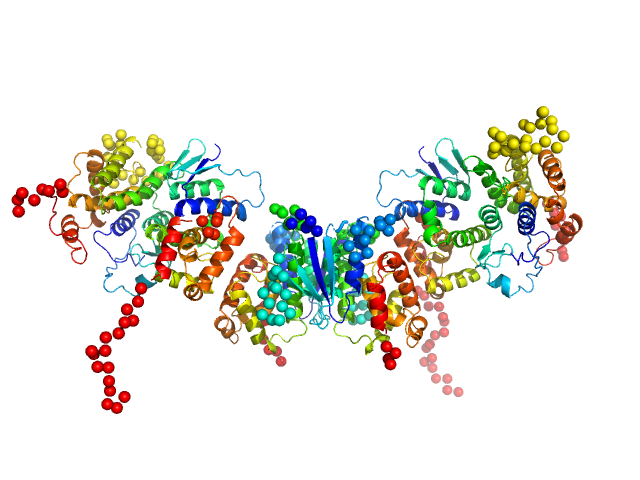
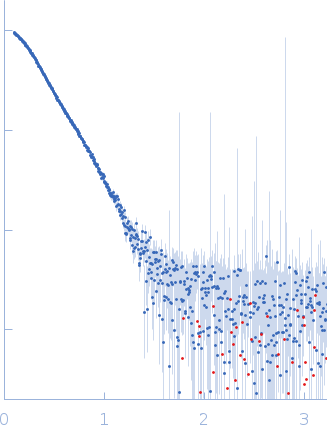
Synchrotron SAXS data of the P. berghei multi-aminoacyl-tRNA synthetase M complex containing the N-terminal GST-like domains of tRNA import protein (tRip-N), glutamyl-tRNA synthetase (ERS-N) and methionyl-tRNA synthetase (MRS-N). The experiment was performed on the SWING beamline at the SOLEIL synchrotron (Saint-Aubin, France) using an EigerX 4M at a sample-detector distance of 2 m and at a wavelength of λ = 0.1033 nm. In-line size-exclusion chromatography was used to separate the sample prior to X-ray exposure. 50 μl of purified recombinant (E. coli) complex at 9 mg/mL were injected on a Bio SEC-3 column (4.6 × 300 mm, 300 Å) equilibrated in 25 mM HEPES-NaOH pH 7.0, 300 mM NaCl, 5% glycerol, 0.005% (m/v) DDM, 5 mM 2-mercaptoethanol at 0.2 mL/min and 15°C. Frames (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle) were collected successively each 1 s (0.990 s exposure and 0.01 dead time) in two time windows. 180 solvent frames were collected at time 2.5 min prior to the column's void volume and 1230 sample frames were collected from time 9.5 min until the end of the run. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent was subtracted from the sample frames using Foxtrot 3.5.10. Data analysis was performed with BioXTAS RAW 2.1.0. The experimental molecular weight was determined using the method of the volume of correlation. A correction factor has been applied to the Porod volume. The atomic model of tRip-N was built with MODELLER 10.1 based on the crystal structure of P. vivax tRip-N (PDB 5ZKF). The crystal structure of ERS-N is available (PDB 8BCQ). MRS-N is a prediction model obtained with Raptor X. The association of the different domains was predicted with ColabFold. The derived model of the M-complex provided an excellent fit to the SAXS data and was consistent with our previous mutagenesis experiments. A dimer of ERS-N (GST-like interface 1) binds two molecules of tRip-N from each side (GST-like interface 2), which in turn associate with two monomers of MRS-N (GST-like interface 1).
|
|
 s, nm-1
s, nm-1