|
X-ray synchrotron radiation scattering data from solutions of periplasmic holdase chaperone protein Skp in 25 mM HEPES 150 mM NaCl, 1 mM DTT, pH 7.5 were measured at the EMBL-P12 bioSAXS beam line on the PETRAIII storage ring (Hamburg, Germany) using a Pilatus 2M detector ((I(s) vs s, where s = 4π sin θ/λ; 2θ is the scattering angle; λ = 0.12 nm). Different solute concentrations in the range 0.58-5.82 mg/ml were measured where each sample and corresponding matched solvent blank were collected as 20 successive 50 ms frames for a total exposure time of 1 s. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted and the different curves were scaled for protein concentration. The low angle data collected from the 0.58 mg/ml sample were merged with the highest concentration (5.82 mg/ml) high angle data to yield the final composite scattering curve. The original SAXS data used to obtain the merged profile as displayed in this entry are included in the full entry zip archive.
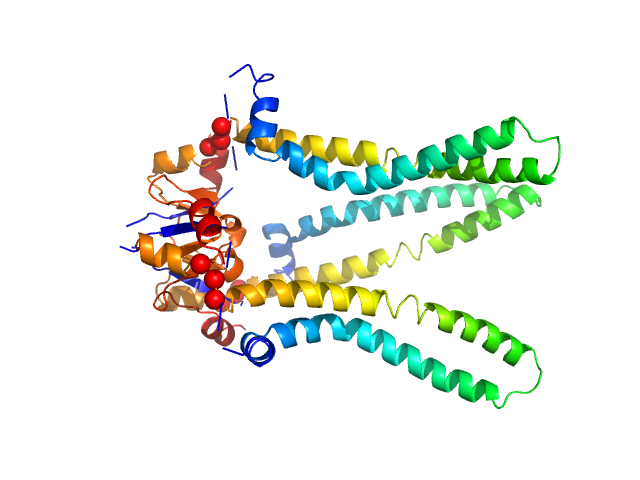
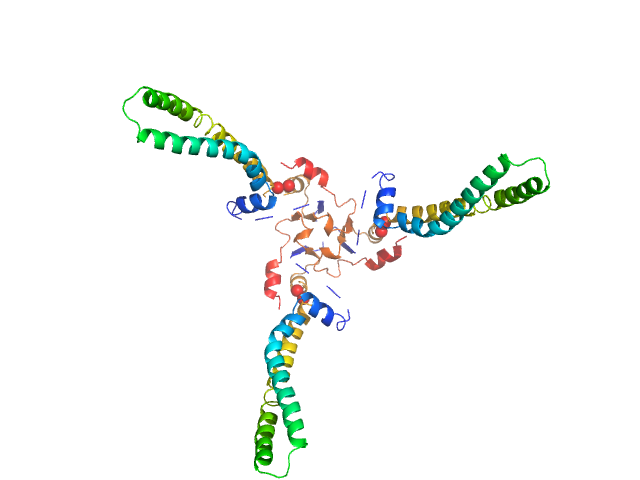
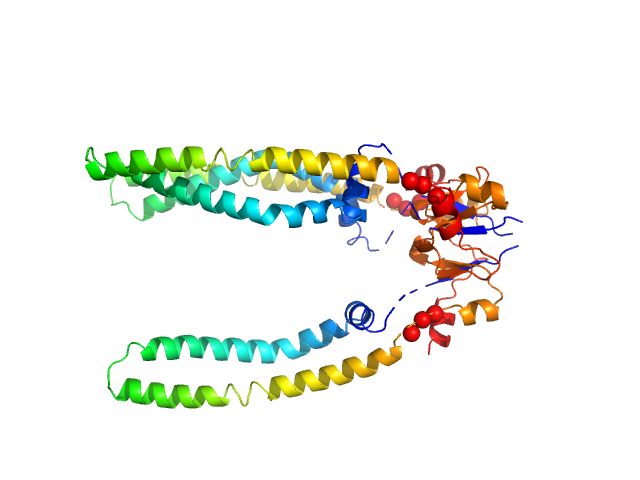
The models presented above are representatives from the trimeric ensemble state(s) of Skp as determined using Ensemble Optimization Method (EOM). The Rg and size distributions obtained from EOM modelling are included in the full entry zip archive.
|
|
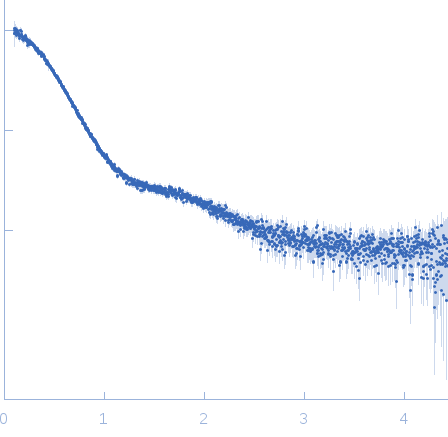 s, nm-1
s, nm-1
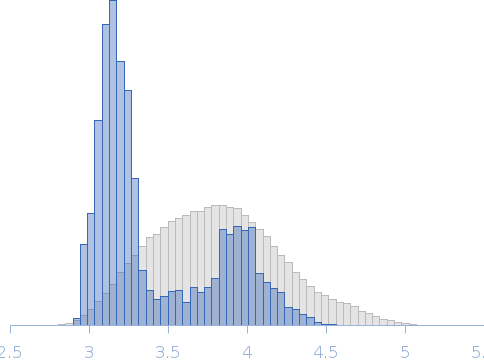 Rg, nm
Rg, nm