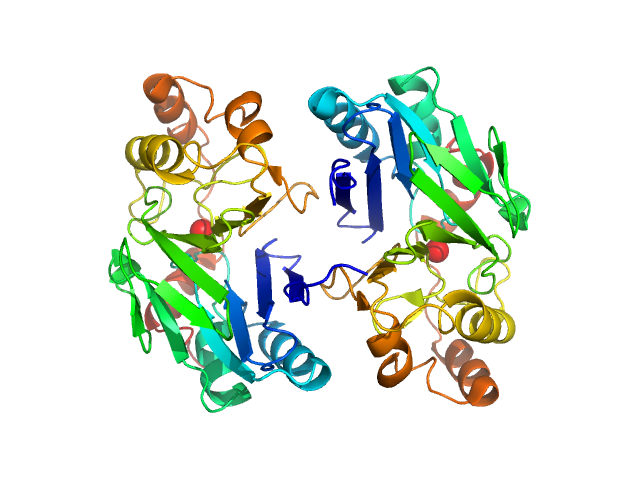
UniProt ID: O95571 (None-None) Persulfide dioxygenase ETHE1, mitochondrial
|
|
|
|
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 21
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
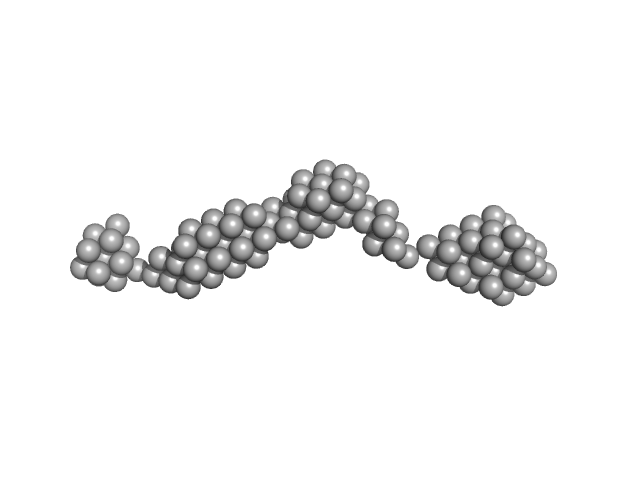
UniProt ID: O95571 (None-None) Persulfide dioxygenase ETHE1, mitochondrial
|
|
|
|
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 23
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
9.0 |
nm |
| Dmax |
32.3 |
nm |
| VolumePorod |
400 |
nm3 |
|
|
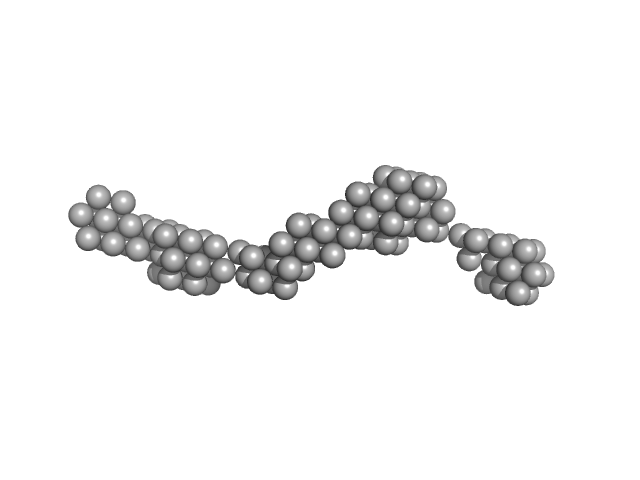
UniProt ID: O95571 (None-None) Persulfide dioxygenase ETHE1, mitochondrial
|
|
|
|
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 23
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
8.3 |
nm |
| Dmax |
38.0 |
nm |
| VolumePorod |
602 |
nm3 |
|
|
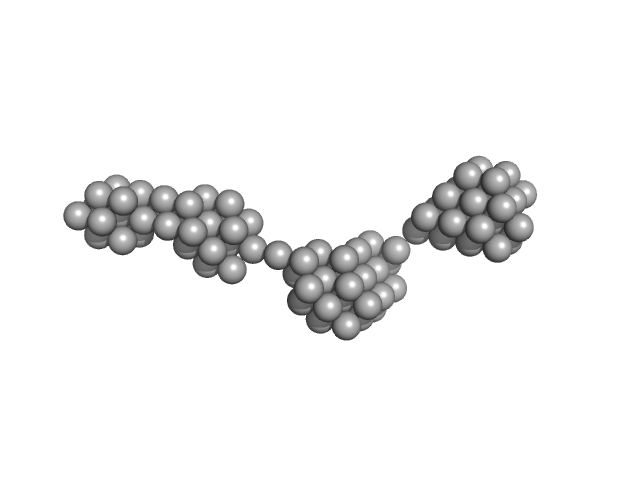
UniProt ID: O95571 (None-None) Persulfide dioxygenase ETHE1, mitochondrial
|
|
|
|
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 23
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
8.8 |
nm |
| Dmax |
42.5 |
nm |
| VolumePorod |
647 |
nm3 |
|
|
UniProt ID: O95571 (None-None) Persulfide dioxygenase ETHE1, mitochondrial
|
|
|

|
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 23
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
11.2 |
nm |
| Dmax |
56.0 |
nm |
| VolumePorod |
1445 |
nm3 |
|
|
UniProt ID: O95571 (None-None) Persulfide dioxygenase ETHE1, mitochondrial
|
|
|
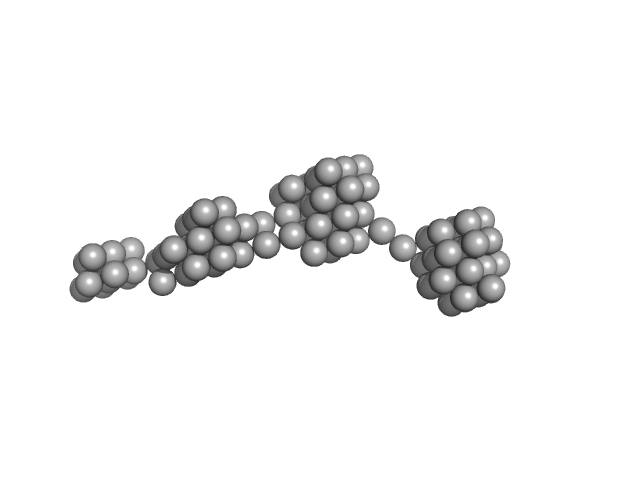
|
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 23
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
12.4 |
nm |
| Dmax |
59.0 |
nm |
| VolumePorod |
1820 |
nm3 |
|
|
UniProt ID: O95571 (None-None) Persulfide dioxygenase ETHE1, mitochondrial
|
|
|
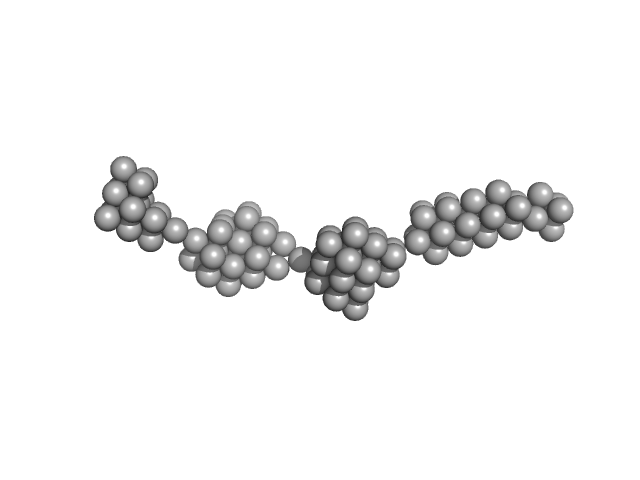
|
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 23
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
14.2 |
nm |
| Dmax |
63.5 |
nm |
| VolumePorod |
2317 |
nm3 |
|
|