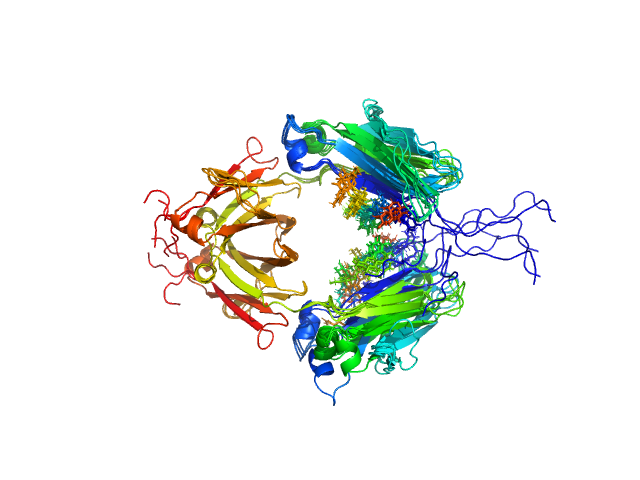
UniProt ID: P01857 (None-None) Immunoglobulin heavy constant gamma 1
|
|
|
|
| Sample: |
Immunoglobulin heavy constant gamma 1 dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 50mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Feb 17
|
Conformational Plasticity of the Immunoglobulin Fc Domain in Solution.
Structure 26(7):1007-1014.e2 (2018)
Remesh SG, Armstrong AA, Mahan AD, Luo J, Hammel M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
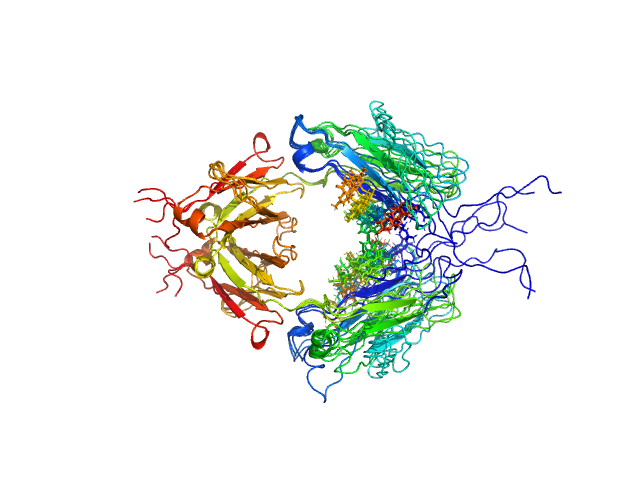
UniProt ID: P01857 (None-None) Immunoglobulin heavy constant gamma 1 M255Y/S257T/T259E
|
|
|
|
| Sample: |
Immunoglobulin heavy constant gamma 1 M255Y/S257T/T259E dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 50mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Feb 17
|
Conformational Plasticity of the Immunoglobulin Fc Domain in Solution.
Structure 26(7):1007-1014.e2 (2018)
Remesh SG, Armstrong AA, Mahan AD, Luo J, Hammel M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
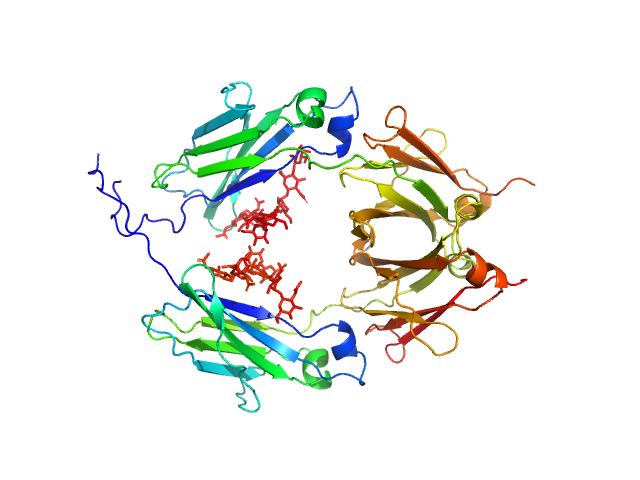
UniProt ID: P01857 (108-329) Glycosylated human immunoglobulin G Fc region
|
|
|
|
| Sample: |
Glycosylated human immunoglobulin G Fc region dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM Citrate-Phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Mar 5
|
CH2 domain orientation of human immunoglobulin G in solution: Structural comparison of glycosylated and aglycosylated Fc regions using small-angle X-ray scattering.
MAbs (2018)
Yageta S, Imamura H, Shibuya R, Honda S
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
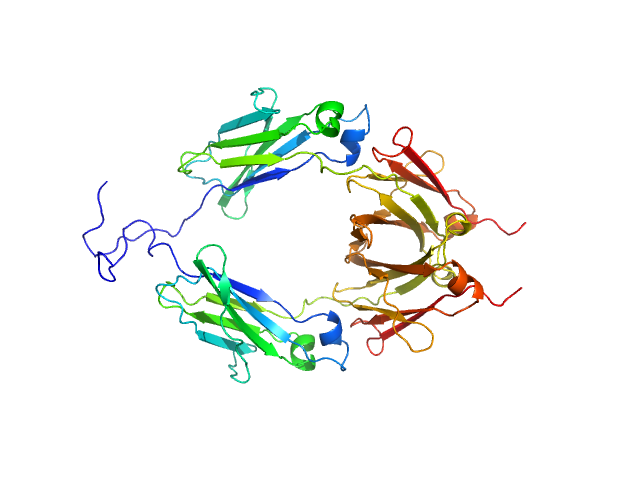
UniProt ID: P01857 (104-330) Aglycosylated human immunoglobulin G Fc region
|
|
|
|
| Sample: |
Aglycosylated human immunoglobulin G Fc region dimer, 51 kDa Homo sapiens protein
|
| Buffer: |
20 mM Citrate-Phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Mar 5
|
CH2 domain orientation of human immunoglobulin G in solution: Structural comparison of glycosylated and aglycosylated Fc regions using small-angle X-ray scattering.
MAbs (2018)
Yageta S, Imamura H, Shibuya R, Honda S
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
UniProt ID: None (None-None) Immunoglobulin heavy constant gamma 1
|
|
|
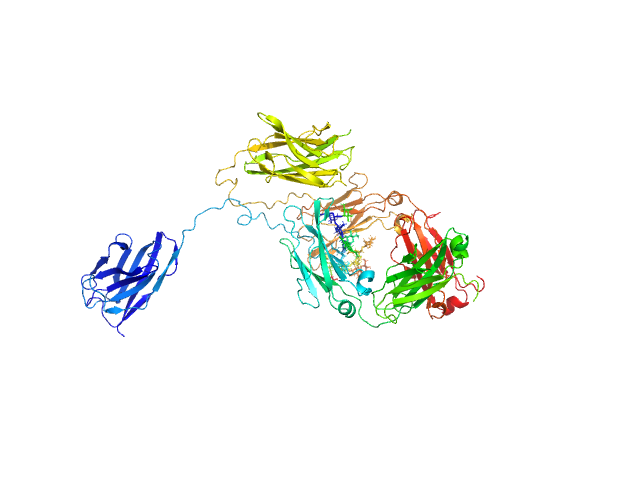
|
| Sample: |
Immunoglobulin heavy constant gamma 1 monomer, 78 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.6 mM KCl buffer, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Sep 28
|
The solution structure of the heavy chain-only C5-Fc nanobody reveals exposed variable regions that are optimal for COVID-19 antigen interactions.
J Biol Chem :105337 (2023)
Gao X, Thrush JW, Gor J, Naismith JH, Owens RJ, Perkins SJ
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
157 |
nm3 |
|
|
UniProt ID: None (None-None) Immunoglobulin heavy constant gamma 1
|
|
|
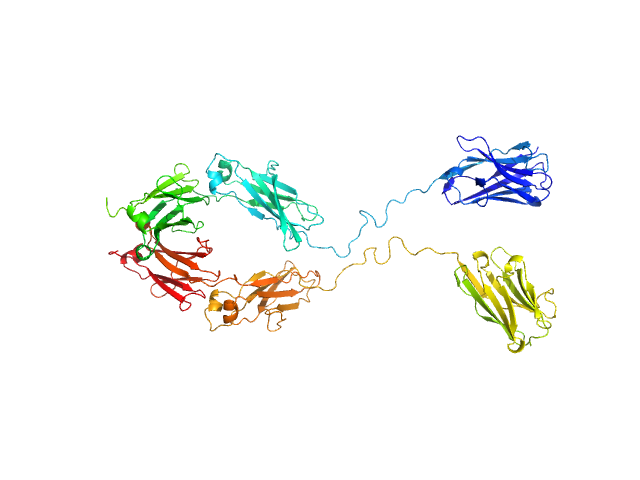
|
| Sample: |
Immunoglobulin heavy constant gamma 1 monomer, 78 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.6 mM KCl buffer, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Sep 28
|
The solution structure of the heavy chain-only C5-Fc nanobody reveals exposed variable regions that are optimal for COVID-19 antigen interactions.
J Biol Chem :105337 (2023)
Gao X, Thrush JW, Gor J, Naismith JH, Owens RJ, Perkins SJ
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.1 |
nm |
| VolumePorod |
175 |
nm3 |
|
|