UniProt ID: P02768 (None-None) Albumin
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.5 |
nm |
|
|
UniProt ID: P01308 (25-110) Insulin detemir (Levemir(R), Novo Nordisk A/S)
UniProt ID: P02768 (25-609) Human Albumin (Recombumin(R) Alpha, Albumedix Ltd.)
|
|
|
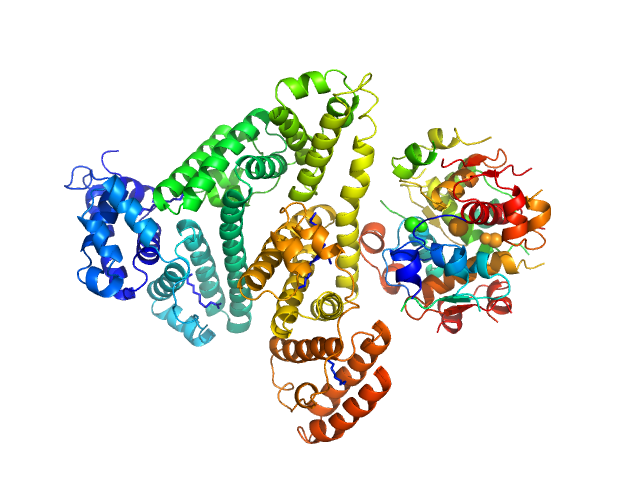
|
| Sample: |
Insulin detemir (Levemir(R), Novo Nordisk A/S) hexamer, 35 kDa protein
Human Albumin (Recombumin(R) Alpha, Albumedix Ltd.) monomer, 66 kDa protein
|
| Buffer: |
6.9 mM Na2HPO4, 11.9 mM m-cresol, 13.7 mM phenol, 157.3 mM glycerol, 38.5 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2015 Sep 25
|
Solution structures of long-acting insulin analogues and their complexes with albumin.
Acta Crystallogr D Struct Biol 75(Pt 3):272-282 (2019)
Ryberg LA, Sønderby P, Barrientos F, Bukrinski JT, Peters GHJ, Harris P
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
162 |
nm3 |
|
|
UniProt ID: P02768 (None-None) Albumin
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.2 |
nm |
|
|
UniProt ID: P01308 (25-110) Insulin detemir (Levemir(R), Novo Nordisk A/S)
UniProt ID: P02768 (25-609) Human Albumin (Recombumin(R) Alpha, Albumedix Ltd.)
|
|
|
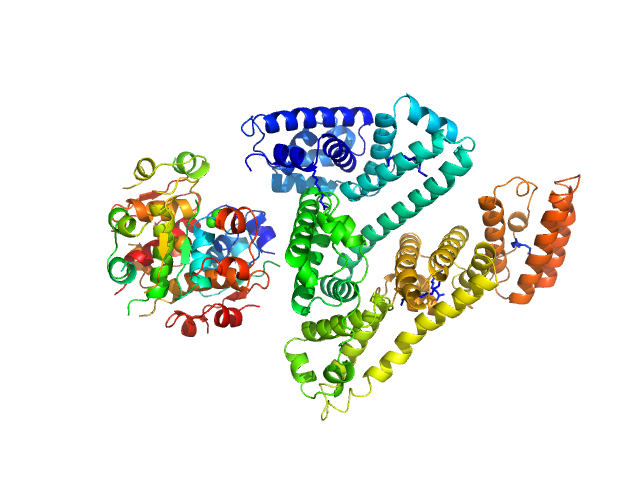
|
| Sample: |
Insulin detemir (Levemir(R), Novo Nordisk A/S) hexamer, 35 kDa protein
Human Albumin (Recombumin(R) Alpha, Albumedix Ltd.) monomer, 66 kDa protein
|
| Buffer: |
6.9 mM Na2HPO4, 11.9 mM m-cresol, 13.7 mM phenol, 157.3 mM glycerol, 38.5 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2015 Sep 25
|
Solution structures of long-acting insulin analogues and their complexes with albumin.
Acta Crystallogr D Struct Biol 75(Pt 3):272-282 (2019)
Ryberg LA, Sønderby P, Barrientos F, Bukrinski JT, Peters GHJ, Harris P
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
162 |
nm3 |
|
|
UniProt ID: P02768 (None-None) Albumin
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
8.9 |
nm |
|
|
UniProt ID: P02768 (25-609) Human Albumin (Recombumin(R) Alpha, Albumedix Ltd.)
UniProt ID: P01308 (None-None) Insulin detemir (Levemir(R), Novo Nordisk A/S)
|
|
|
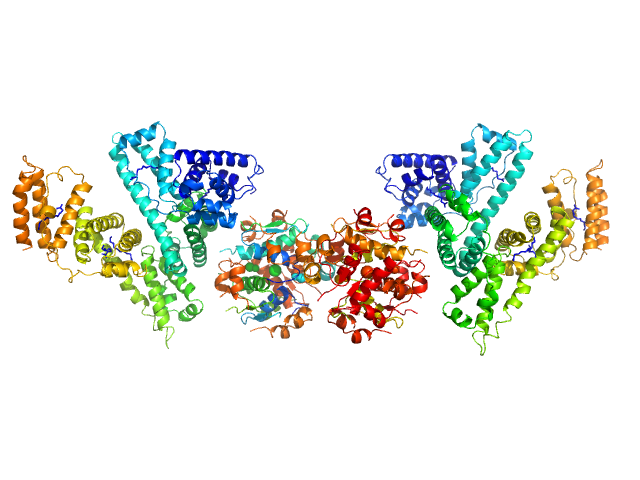
|
| Sample: |
Human Albumin (Recombumin(R) Alpha, Albumedix Ltd.) monomer, 66 kDa protein
Insulin detemir (Levemir(R), Novo Nordisk A/S) dodecamer, 71 kDa protein
|
| Buffer: |
8.8 mM Na2HPO4, 10.6 mM m-cresol, 12.2 mM phenol, 140.9 mM glycerol, 56.9 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2015 Sep 25
|
Solution structures of long-acting insulin analogues and their complexes with albumin.
Acta Crystallogr D Struct Biol 75(Pt 3):272-282 (2019)
Ryberg LA, Sønderby P, Barrientos F, Bukrinski JT, Peters GHJ, Harris P
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
309 |
nm3 |
|
|
UniProt ID: P02768 (None-None) Albumin
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.1 |
nm |
|
|
UniProt ID: P02768 (25-609) Human Albumin (Recombumin(R) Alpha, Albumedix Ltd.)
UniProt ID: P01308 (None-None) Insulin detemir (Levemir(R), Novo Nordisk A/S)
|
|
|
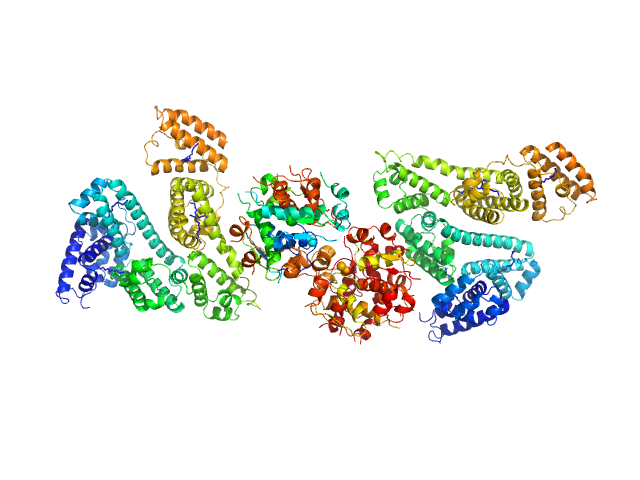
|
| Sample: |
Human Albumin (Recombumin(R) Alpha, Albumedix Ltd.) monomer, 66 kDa protein
Insulin detemir (Levemir(R), Novo Nordisk A/S) dodecamer, 71 kDa protein
|
| Buffer: |
8.8 mM Na2HPO4, 10.6 mM m-cresol, 12.2 mM phenol, 140.9 mM glycerol, 56.9 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2015 Sep 25
|
Solution structures of long-acting insulin analogues and their complexes with albumin.
Acta Crystallogr D Struct Biol 75(Pt 3):272-282 (2019)
Ryberg LA, Sønderby P, Barrientos F, Bukrinski JT, Peters GHJ, Harris P
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
309 |
nm3 |
|
|
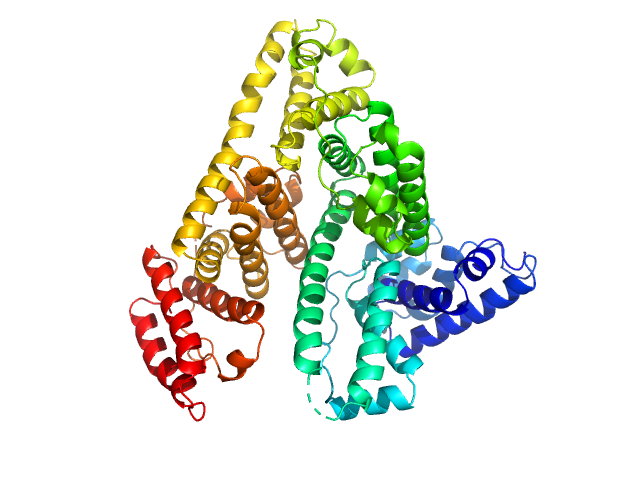
UniProt ID: P02768 (None-None) Albumin
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.2 |
nm |
|
|
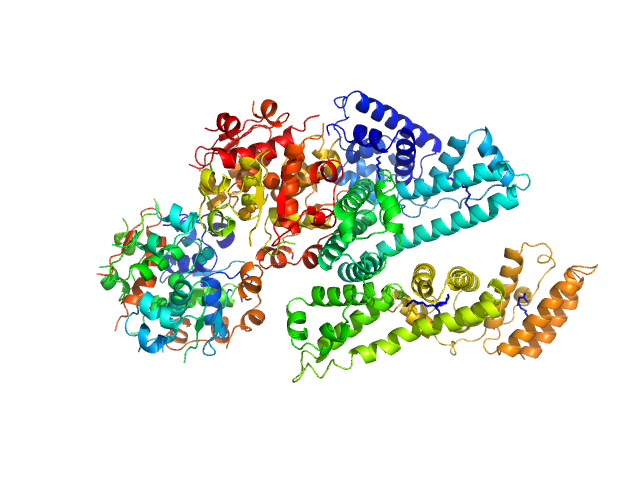
UniProt ID: P02768 (25-609) Human Albumin (Recombumin(R) Elite, Albumedix Ltd.)
UniProt ID: P01308 (25-110) Insulin degludec(Tresiba(R), Novo Nordisk A/S)
|
|
|
|
| Sample: |
Human Albumin (Recombumin(R) Elite, Albumedix Ltd.) monomer, 66 kDa protein
Insulin degludec(Tresiba(R), Novo Nordisk A/S) dodecamer, 73 kDa protein
|
| Buffer: |
25 mM Na2HPO4, 15.9 mM m-cresol, 15.9 mM phenol, 212.8 mM glycerol, 20 mM NaCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 27
|
Solution structures of long-acting insulin analogues and their complexes with albumin.
Acta Crystallogr D Struct Biol 75(Pt 3):272-282 (2019)
Ryberg LA, Sønderby P, Barrientos F, Bukrinski JT, Peters GHJ, Harris P
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
179 |
nm3 |
|
|