UniProt ID: P0DP23 (1-149) Calmodulin-1
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 4 mM CaCl₂, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Sep 24
|
A High‐Affinity Calmodulin‐Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells
Advanced Science :2003630 (2021)
Voegele A, Sadi M, O'Brien D, Gehan P, Raoux‐Barbot D, Davi M, Hoos S, Brûlé S, Raynal B, Weber P, Mechaly A, Haouz A, Rodriguez N, Vachette P, Durand D, Brier S, Ladant D, Chenal A
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
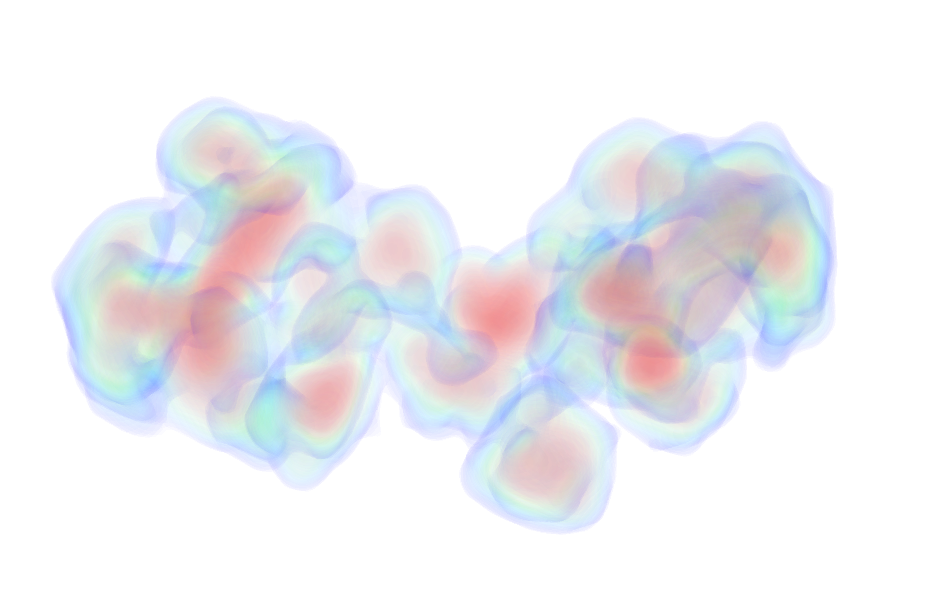
UniProt ID: P0DP23 (1-149) Calmodulin-1
UniProt ID: P0DKX7 (454-484) Bifunctional hemolysin/adenylate cyclase
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
Bifunctional hemolysin/adenylate cyclase monomer, 3 kDa Bordetella pertussis protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 4 mM CaCl₂, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Sep 24
|
A High‐Affinity Calmodulin‐Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells
Advanced Science :2003630 (2021)
Voegele A, Sadi M, O'Brien D, Gehan P, Raoux‐Barbot D, Davi M, Hoos S, Brûlé S, Raynal B, Weber P, Mechaly A, Haouz A, Rodriguez N, Vachette P, Durand D, Brier S, Ladant D, Chenal A
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
UniProt ID: P0DP23 (2-149) Calmodulin-1
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 2 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Nov 15
|
Dynamics and structural changes of calmodulin upon interaction with the antagonist calmidazolium.
BMC Biol 20(1):176 (2022)
Léger C, Pitard I, Sadi M, Carvalho N, Brier S, Mechaly A, Raoux-Barbot D, Davi M, Hoos S, Weber P, Vachette P, Durand D, Haouz A, Guijarro JI, Ladant D, Chenal A
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
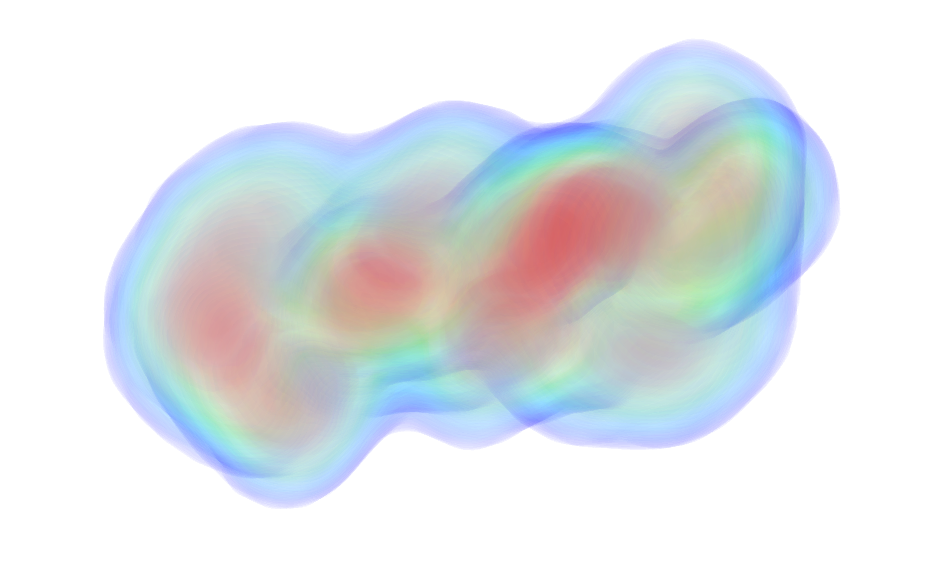
UniProt ID: P0DP23 (2-149) Calmodulin-1
UniProt ID: None (None-None) Calmidazolium
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
Calmidazolium monomer, 1 kDa
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 2 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Nov 15
|
Dynamics and structural changes of calmodulin upon interaction with the antagonist calmidazolium.
BMC Biol 20(1):176 (2022)
Léger C, Pitard I, Sadi M, Carvalho N, Brier S, Mechaly A, Raoux-Barbot D, Davi M, Hoos S, Weber P, Vachette P, Durand D, Haouz A, Guijarro JI, Ladant D, Chenal A
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.2 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
UniProt ID: P0DP23 (1-149) Calmodulin-1
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 2 mM CaCl2, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Sep 23
|
Binding by calmodulin is coupled to transient unfolding of the third FF domain of Prp40A.
Protein Sci 32(4):e4606 (2023)
Díaz Casas A, Cordoba JJ, Ferrer BJ, Balakrishnan S, Wurm JE, Pastrana-Ríos B, Chazin WJ
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
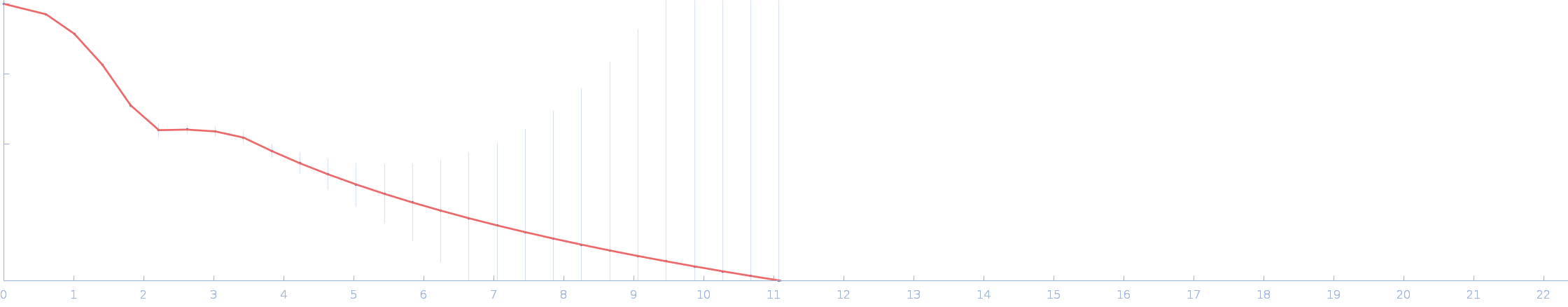
UniProt ID: P0DP23 (1-149) Calmodulin-1
UniProt ID: O75400 (516-592) Pre-mRNA-processing factor 40 homolog A
|
|
|
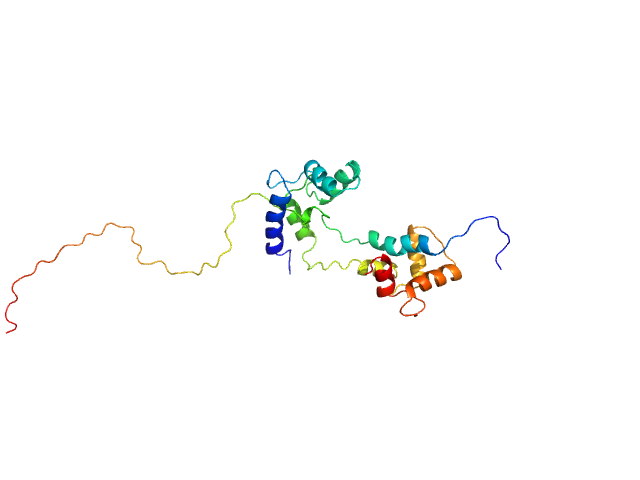
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
Pre-mRNA-processing factor 40 homolog A monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 2 mM CaCl2, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Sep 23
|
Binding by calmodulin is coupled to transient unfolding of the third FF domain of Prp40A.
Protein Sci 32(4):e4606 (2023)
Díaz Casas A, Cordoba JJ, Ferrer BJ, Balakrishnan S, Wurm JE, Pastrana-Ríos B, Chazin WJ
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
UniProt ID: P0DP23 (1-149) Calmodulin-1
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Jun 4
|
New insights into P2X7 receptor regulation: Ca2+-calmodulin and GDP bind to the soluble P2X7 ballast domain.
J Biol Chem :102495 (2022)
Sander S, Müller I, Alai MG, Nicke A, Tidow H
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
UniProt ID: P0DP23 (1-149) Calmodulin-1
UniProt ID: Q99572 (395-595) P2X purinoceptor 7
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
P2X purinoceptor 7 monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Jun 4
|
New insights into P2X7 receptor regulation: Ca2+-calmodulin and GDP bind to the soluble P2X7 ballast domain.
J Biol Chem :102495 (2022)
Sander S, Müller I, Alai MG, Nicke A, Tidow H
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
73 |
nm3 |
|
|